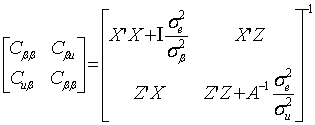ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2006; 2(4):171-178. doi:10.7150/ijbs.2.171 This issue Cite
Research Paper
A novel nuclear-encoded mitochondrial poly(A) polymerase PAPD1 is a potential candidate gene for the extreme obesity related phenotypes in mammals
1. Department of Animal Sciences, Washington State University, Pullman, WA 99164- 6351, USA
2. School of Biological Sciences, Washington State University, Pullman, WA 99164- 4236 USA
Abstract
People with obesity, especially extreme obesity, are at risk for many health problems. However, the responsible genes remain unknown in >95% of severe obesity cases. Our previous genome-wide scan of Wagyu x Limousin F2 cattle crosses with extreme phenotypes revealed a molecular marker significantly associated with intramuscular fat deposition. Characterization of this marker showed that it is orthologous to the human gene KIAA1462 located on HSA10p11.23, where a major quantitative trait locus for morbid obesity has been reported. The newly identified mitochondrial poly(A) polymerase associated domain containing 1 (PAPD1) gene, which is located near this marker, is particularly interesting because the polymerase is required for the polyadenylation and stabilization of mammalian mitochondrial mRNAs. In the present study, both cDNA and genomic DNA sequences were annotated for the bovine PAPD1 gene and ten genetic markers were detected in the promoter and exon 1 region. Among seven markers assayed on ~ 250 Wagyu x Limousin F2 animals, two single nucleotide polymorphisms (SNPs) in the promoter region were significantly associated with intramuscular fat (P<0.05). However, there was a significant interaction (P<0.05) between a third SNP, which causes an amino acid change in coding exon 1, and each of these two promoter SNPs on intramuscular fat deposition. In particular, the differences between double heterozygous animals at two polymorphic sites and the slim genotype animals exceeded 2.3 standard deviations for the trait in both cases. Our study provides evidence for a new mechanism – the involvement of compound heterosis in extreme obesity, which warrants further examination.
Keywords: PAPD1, Nuclear-encoded mitochondrial gene, Polymorphisms, Compound heterosis, Extreme obesity.
1. Introduction
Obesity has increased at a fast rate over the past two decades and is now a worldwide leading public health problem. In 1991, only four of 45 participating states in the United States had obesity prevalence rates of 15 to 19% and none had prevalence greater than 20% [1]. By the year 2004, however, seven of 49 participating states had obesity prevalence rates of 15–19 percent, 33 states had rates of 20–24 percent and 9 states had rates more than 25 percent (http://www.cdc.gov/). Overall, more than 65% of U.S. adults are overweight, with nearly 31% (more than 61 million people) meeting the criteria for obesity. Furthermore, the greatest increases have been observed in the prevalence of extreme obesity [2]. Unfortunately, although over 600 genes, markers and chromosomal regions have been identified as associated with or linked to human obesity phenotypes [3], the responsible genes are still unknown in >95% of severe obesity cases [4].
Recently, we used an amplified fragment length polymorphism (AFLP) approach to screen genome regions for extreme obesity-related phenotypes on divergent animals derived from Wagyu x Limousin crosses. The Wagyu breed of cattle has been traditionally selected for high intramuscular fat (average marbling score greater than 8.52), whereas the Limousin breed has been selected for heavy muscle, which leads to low intramuscular fat (average marbling score less than 4.78) [5]. The difference in the trait between these two breeds makes them very unique for mapping genes for obesity-related phenotypes. Characterization of one of the AFLP fragments that was significantly different in frequency between animals with extreme high or low intramuscular fat content or marbling indicated that it is orthologous to the human gene KIAA1462 located on HSA10p11.23. Several studies on different populations have shown that this human chromosomal region harbors a major quantitative trait locus linked to or associated with morbid obesity [6-8]. The gene PAPD1, poly(A) polymerase associated domain containing 1 is just adjacent to the human gene KIAA1462 on the chromosome. It is a newly discovered nuclear-encoded mitochondrial poly(A) polymerase that has a role in mitochondrial RNA processing [9]. The polymerase is required for the polyadenylation and stabilization of mammalian mitochondrial mRNAs [10]. In-silico mapping could place the bovine PAPD1 gene to a region of 45.589 and 46.63 cM on bovine chromosome 13.
Mitochondria perform a large number of reactions in mammalian cells. In particular, the major site of fatty acid ß-oxidation occurs in mitochondria [11], which may provide key intermediates for the synthesis of triglycerides via the action of pyruvate carboxylase [12]. Therefore, genes that are involved in regulating and stimulating mitochondrial biogenesis would affect the key aspects of adipogenesis and consequently contribute to development of the obesity-related phenotypes in mammals. Wilson-Fritch and colleagues [13] found a profound decrease of approximately 50% in the levels of transcripts for nuclear-encoded mitochondrial genes accompanying the onset of obesity. Recently, Jiang and colleagues [14] found that bovine mitochondrial transcription factor A (TFAM) was significantly associated with intramuscular fat (marbling) and subcutaneous fat depth in the same population of cattle as described above. Here we report for the first time that PAPD1 contributes significantly to the extreme obesity phenotypes, perhaps due to a compound heterosis.
2. Materials and Methods
Compilation of cDNA and genomic DNA sequences for the bovine PAPD1 gene
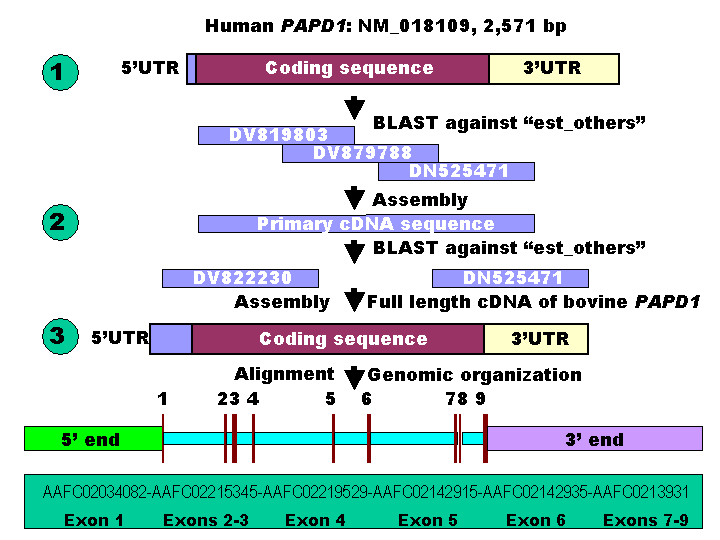
A three-step bioinformatics procedure was used for retrieving both cDNA and genomic DNA sequences of the bovine PAPD1 gene in the present study (Figure 1). In step 1, the cDNA sequence of the human PAPD1 gene was used as a reference to retrieve the orthologous ESTs (expressed sequence tags) against the GenBank database “est_others” with a species option limited to Bos taurus. In step 2, several ESTs were chosen and assembled to form a primary cDNA sequence for the bovine gene, which was then used to perform a species-specific ESTs search against the same database in order to expand the primary sequence to a full-length cDNA sequence. Finally, the full-length cDNA sequence was used to search for genomic DNA sequences against the 6X bovine genome sequence database (http://www.hgsc.bcm.tmc.edu/projects/bovine/). Alignment of the full-length cDNA sequence with the genomic DNA sequence determined its genomic organization. The ORF (open reading frame) finder developed by the National Center for Biotechnology Information (NCBI) was used to find all open reading frames and deduce amino acid sequence based on the full-length cDNA sequence of the bovine PAPD1 gene.
Comparative annotation of both cDNA and genomic DNA sequences of the bovine PAPD1 gene.
Primer design for amplification of the bovine PAPD1 gene
Three pairs of primers were designed to detect genetic polymorphisms in the bovine PAPD1 gene, based on the genomic sequence generated from the bovine genome sequencing project. The first primer pair (forward sequence, 5'-GAG TGT GGT GGT TAG GGG TGG TAG-3' and reverse sequence, 5'-TTC ACA GTA GGG TTT CCC TTC CTC-3') targeted a 517 bp region of the promoter from bases 9,048 to 9,564 (GenBank accession no. AAFC02034082). The second primer pair (forward sequence, 5'-CCC CGT CTC TGG TTC TAT TTT CAA T 3' and reverse sequence, 5' GCG ACT CCA GAC TCT TCC TCC TG 3') amplified a 481 bp product, which included the entire exon 1 plus part of the promoter sequence, 5'UTR and partial intron 1 sequence (GenBank accession no. AAFC02034082). The last primer pair (forward sequence, 5'-TTC ATT GAG TTA GAC AAG GCT GTG-3' and reverse sequence, 5'-TAG CTC CAA TAC GTG TCA ATT TTT-3') was based on GenBank accession no. AAFC02139310 and amplified a 543 bp product, including partial intron 6, entire exon 7, complete intron 7, entire exon 8 and partial intron 8 sequences.
Detection of genetic polymorphisms in the bovine PAPD1 gene
The animals used in the present research were derived from a Wagyu x Limousin cross, including 6 F1 bulls, 113 F1 dams and ~250 F2 progeny. Two obesity-related phenotypes included beef marbling score (BMS) and subcutaneous fat depth (SFD) were compared to PAPD1 genotypes. Beef marbling score is a subjective measure of the amount of intramuscular fat in the longissimus muscle based on USDA standards, while SFD was measured at the 12-13th rib interface perpendicular to the outside surface at a point three-fourths the length of the longissimus muscle from its chine bone end. Genetic polymorphisms were screened by comparison of sequence patterns on six F1 bulls. The PCR reactions were performed in a total volume of 10 µl, including 25 ng of bovine genomic DNA, 1X Platinum Taq Buffer (20 mM Tris-HCl, pH 8.4, 50 mM KCl; Invitrogen), 3.0 mM MgCl2, 0.3 mM each of the four dNTPs, 0.5 U Platinum Taq polymerase (Invitrogen) and 25 ng of each primer. The PCR reactions started with denaturation at 94°C for 2 min, followed by 8 touch-down cycles: 30 seconds at 94°C, 30 seconds at 71°C to 63°C (i.e. decreasing 1°C per cycle), and 30 seconds at 72°C. Then, the PCR took another 37 cycles of reactions: 30 seconds at 94°C, 30 seconds at 63°C and 30 seconds at 72°C, and finally ended after 5 min at 72°C. The PCR products were then sequenced on an ABI 3730 sequencer in the Laboratory for Biotechnology and Bioanalysis at Washington State University following a standard protocol.
Assay development for marker genotyping in the bovine PAPD1 gene
Six single nucleotide polymorphisms (SNPs) were detected in the promoter region, which were then genotyped individually using a direct PCR sequencing approach. Four SNPs were found in the exon 1 region, including two coding SNPs and two genomic SNPs, respectively. Of these two coding SNPs, one is a silent mutation and the other is a missense mutation. The missense mutation could be genotyped using a PCR-RFLP (restriction fragment length polymorphism) approach. Therefore, the PCR amplicons were digested at 37°C for three hours with 2U of StuI for the G/A SNP, followed by analysis on 1.6% agarose gels and visualized by staining with ethidium bromide. However, no polymorphisms were detected in the product spanning exon 7 and exon 8. Therefore, among the ten SNPs detected in the bovine PAPD1 gene, seven were successfully assayed on the reference population.
Association of the bovine PAPD1 gene with the obesity-related phenotypes
The phenotypic data have been previously adjusted for year of birth, sex, age (days), live weight (kilograms), or fat depth (centimeters), as appropriate. The adjusted phenotypes were then used in a subsequent association analysis based on the following mixed model:
y=Xβ+Zu+e (1)
where y was a vector containing all observations, β was a vector for fixed effects including marker all genotypes or genotype combinations, u ~ N(0, Aσu2 ) was a vector for residual genetic effects that were not accounted for by the genes (markers) under investigation, A was a numerator relationship matrix, X and Z were incidence matrices which link observations in y to effects in β and u, respectively, and e ~ N(0, Iσe2 ) was a vector for residual errors. Model parameters were estimated using REML (residual maximum likelihood) and Bayesian analysis, respectively. In the Bayesian estimation, we assumed normal priors for fixed effects and chi-square distributions for the variance components, σu2 and σe2, respectively. Markov Chain Monte Carlo (MCMC) was used to simulate model parameters based on their fully conditional distributions below.
(2)
where:
;
(3)
(4)
Where aii' was the element in row i and column i' of the q×q inverse matrix A-1 , r = y-Xβ-Zu was a vector containing residual errors, vk and Sk2 are the degrees of freedom parameter and the scaling factor in the prior distribution of 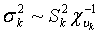 (k = u, e).
(k = u, e).
3. Results
Annotation of the bovine PAPD1 gene
The BLAST search using the cDNA sequence of the human PAPD1 gene (NM_018109) retrieved more than 20 orthologous ESTs in cattle against “est_others” in the GenBank database. Three ESTs - DV819803, DV879788 and DN525471 were used to form a preliminary cDNA sequence of the bovine PAPD1 gene (Figure 1), which was then used as a query for a second BLAST run to retrieve both 5' and 3' cDNA end sequences missed by the first BLAST run using the cDNA sequence of the human gene as a reference. Adding two ESTs (DV822230 and CV974359) with the three ESTs described above contributed to the formation of a full-length cDNA sequence of 2,554 bp for the bovine gene. This newly assembled bovine gene hit more than 30 ESTs in the database with sequence identities of 98 – 100%. ORF finder detected a longest coding sequence of 1,701 bp with a total of 566 deduced amino acids for the bovine PAPD1 protein. The full-length cDNA sequence of the bovine PAPD1 gene retrieved six genomic DNA contigs (AAFC02034082, AAFC02215345, AAFC02219529, AAFC02142915, AAFC02142935 and AAFC02139310) from the 6X bovine genome sequence database with a total of 50,536 bp (Figure 1). Alignment of the cDNA with genomic DNA sequences revealed that this bovine gene contains nine exons and eight introns. Among these eight introns, however, only introns 2, 7 and 8 have completed sequences.
Genetic polymorphisms and genotyping
Three regions of the bovine PAPD1 gene were amplified and sequenced on six F1 bulls individually to screen polymorphisms. Sequencing data showed that the promoter region was highly polymorphic: a total of six genetic polymorphisms were detected in a product of 517 bp, with one mutation every 86 bp of sequence. These six polymorphisms include five SNPs (AAFC02034082.1:g.9224G>A, 9330A>C, 9367G>C, 9419A>C and 9435T>G) and one insertion/deletion (AAFC02034082.1:g.9409[(G)4]+[(G)5], respectively. We decided to genotype all these genetic markers in the promoter region using a direct sequencing PCR product approach. In exon 1 and its flanking regions, four SNPs were detected, including two genomic SNPs (AAFC02034082.1:g.10084T>C and 10228G>T) and two coding SNPs (AAFC02034082.1:c.10326C>T and 10364A>G). The latter coding SNP (AAFC02034082.1:c.10364A>G) changes an amino acid from aspartic acid (GAC) to glycine (GGC) and causes a gain/loss of enzyme cut site for StuI. Therefore, a PCR-RFLP assay was developed to genotype this marker on all individuals. No polymorphisms were detected around exon 7 and exon 8 region of the bovine PAPD1 gene.
Associations of bovine PAPD1 gene with beef marbling and SFD
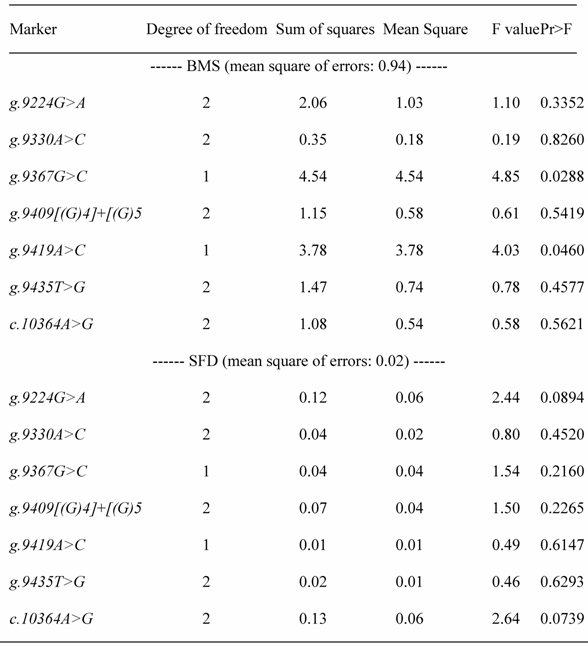
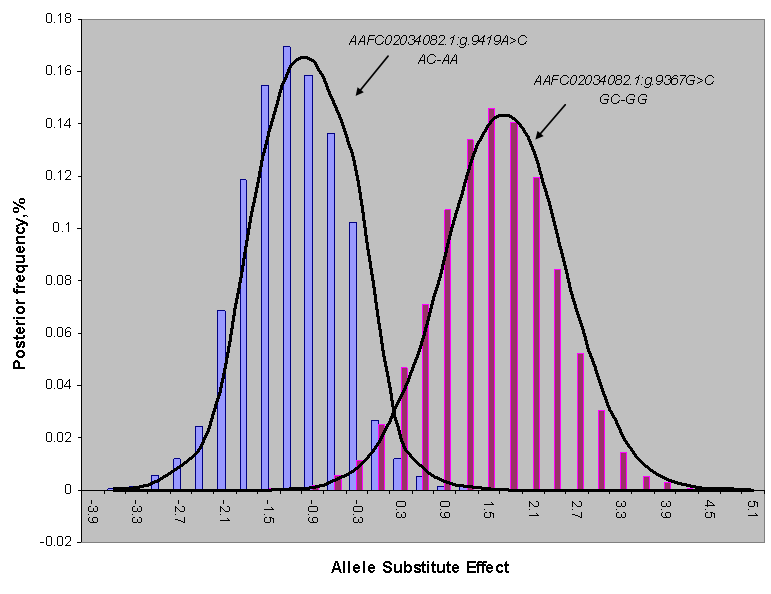
REML-based analysis of variance suggested significant associations of two SNPs in the promoter region (AAFC02034082.1:g.9367 G>C (P=0.0288) and 9419A>C (P=0.0397)) with BMS, and a suggestive association of AAFC02034082.1:c.10364A>G with SFD (P=0.0739) (Table 1). Bayesian analysis was in agreement with the REML-based analysis of variance, yet it gave a more intuitive inference of the association by giving their posterior distribution. For AAFC02034082.1:g.9367G>C, only CG and GG genotypes exist in the F2 progeny. However, for AAFC02034082.1:g.9419A>C, only AC and AA genotypes were considered in the analysis, because only two CC individuals were detected in the F2 progeny. In the Bayesian analysis, over 95% posterior samples showed positive differences between PAPD1:g.9367CG and GG or negative difference between PAPD1:g.9419AC and AA (Figure 2). Therefore, these deviates of means between one heterozygous genotype and one homozygous genotype could be interpreted as allele substitution effects estimated under the assumption of complete linkage between the marker and the causal gene.
Analysis of variance for testing associations between polymorphic markers and beef marbling score (BMS) and subcutaneous fat depth (SFD)
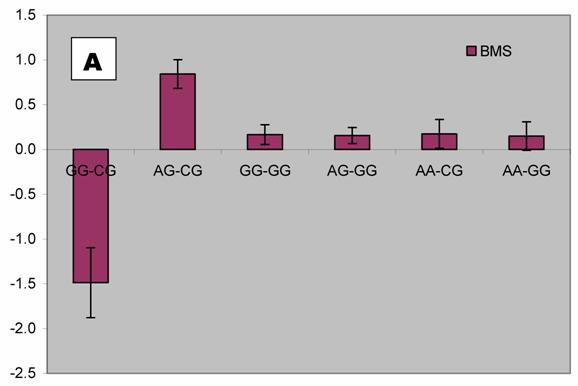
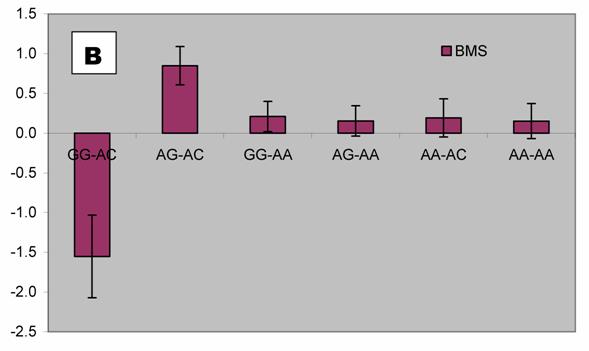
We further analyzed pair-wise interactions between these two markers (AAFC02034082.1:g.9367G>C and 9419A>C), which were significantly associated with BMS, and other markers in the bovine PAPD1 gene. REML analyses indicated that there was a significant interaction between AAFC02034082.1:c.10364A>G and these two markers for beef marbling. Due to the singularity of matrix X'X, interaction effects are not uniquely estimable in the REML analysis. We therefore presented interaction estimates obtained using Bayesian estimation. Interestingly, interactions between two heterozygous genotypes led to the highest marbling scores (Figure 3A and 3B). Animals with both heterozygous genotypes of PAPD1: c.10364AG/g.9367CG had an additional 2.33 and 0.67 - 0.69 marbling scores compared to the genotype combination of PAPD1: c.10364GG/g.9367CG and all other combinations, respectively (P=0.0248) (Figure 3A). The same interaction trend was also observed for markers AAFC02034082.1:c.10364A>G and g.9419A>C (P=0.0055). The marbling score was 2.40 higher in animals with double heterozygote genotypes of PAPD1: c.10364AG/g.9419AC than the genotypes of PAPD1: c.10364GG/g.9419AC. The remaining genotypes at both markers were lower than the double heterozygous genotypes by 0.64 – 0.70 marbling scores (Figure 3B).
Posterior distribution of estimated allele substitution effect of two SNPs in the promoter region (AAFC02034082.1:g.9367G>C and g.9419A>C) on beef marbling score under the assumption of complete linkage between the markers and the causal gene.
Bayesian estimation of interaction effects between markers: A, c.10364A>G and g.9367G>C, and B, c.10364A>G and g.9419A>C, on beef marbling scores (BMS).
4. Discussion
Oxidative phosphorylation is the most important event performed in mitochondria, which transforms the energy of nutrient-derived substrates into the energy stored in ATP [11]. Mitochondria supply the majority of this ATP to the rest of cell. When energy intake exceeds energy expenditure, the resulting imbalance may expand the size and increase the number of fat cells. This process basically leads to fat deposition in humans and in mammals. Therefore, the genes that regulate and stimulate mitochondrial biogenesis would play an important role in adipogenesis [13]. In the present study, we found that the PAPD1 gene, a novel nuclear-encoded mitochondrial poly(A) polymerase contributes to extreme fat deposition in cattle. Evidence has shown that the PAPD1 gene is involved in regulating mitochondrial biogenesis by controlling poly(A) synthesis in human mitochondria [10]. Recently, Jiang and colleagues [14-15] studied the effects of mitochondrial transcription factor A (TFAM), B1 (TFB1M) and B2 (TFB2M) on obesity related phenotypes using the same reference population as described above. Overall, we found that involvement of these four genes, TFAM, TFB1M, TFB2M and PAPD1 in the stimulation and regulation of mitochondrial biogenesis may be tissue-specific or relevant. TFAM contributed significantly to both intramuscular fat deposition and SFD [14], while TFB1M had no effect on either trait. PAPD1 significantly affected intramuscular fat, but not SFD (this study). However, TFB2M contributed more to SFD, but almost nothing to intramuscular fat deposition [15].
In 1998, Hager and colleagues [6] first reported significant evidence for linkage of obesity to a chromosome 10 locus with a maximal logarithm of odds score (MLS) near the D10S197 marker based on a genome-wide scan on 158 multiplex French obese Caucasian families with 514 individuals. This obesity-linked peak at D10S197 on HSA10p11-12 was further confirmed in four other ethnic groups, including 170 European-American and 43 African-American families [8], 89 German families [16] and 188 nuclear families around Paris, France [17]. As D10S197 is located in intron 7 of the GAD2 gene at position 26.54 Mb on HSA10, Boutin and colleagues [17] proposed that the gene GAD2 encoding the glutamic acid decarboxylase enzyme (GAD65) is a positional candidate gene for obesity on the chromosome. Both a case-control study (575 morbidly obese and 646 control subjects) and a familial-based analysis analyzing GAD2 variants confirmed the association of obesity with the SNP +61450 C>A and +83897 T>A haplotype (χ2 = 7.637, p = 0.02). However, fine mapping performed on the same population by the authors revealed a second peak at D10S600 (MLS = 3.4 at this locus vs. MLS = 3.2 at D10S197). The marker D10S600 is located at position 28.70 on HSA10, just 2.16 Mb away from the marker D10S197 (26.54 Mb on HSA10). Recently, Hinney and co-workers [7] further moved the Chromosome 10 obesity peak down to TCF8 with a maximum LOD of 2.32. The authors also found that the two point LOD score was 1.38 only at the marker D10S197. The TCF8 marker is positioned at 31.65 Mb on HSA10. Interestingly, the PAPD1 gene we studied here is located at position 30.64 Mb on HSA10 between both markers D10S600 and TCF8. These data warrant further investigation on the candidacy of the PAPD1 gene as a potential candidate gene for the second obesity peak on HSA10p11-12.
Evidence has shown that heterosis is one of the contributors responsible for the obesity-related phenotypes in mammals. Vaisse and colleagues [18] reported a high frequency (4%) of rare heterozygous MC4-R mutations in a large population of morbidly obese patients, but no such mutations were found in controls. Interestingly, HTR2C promoter variation was found to be a risk factor for obesity and, perhaps through heterosis, influence weight loss in obese women [19]. Among ninety-five obese women who participated in a randomized trial of psychological treatments for weight loss, heterozygotes lost less weight during the trial than did homozygotes (6.8 vs. 9.7 kg; P = 0.047) and weighed more six months (90.1 vs. 83.6 kg; P = 0.006) and 12 months (91.8 vs. 84.6 kg; P = 0.009) later. Heterozygotes also had higher triglyceride levels than homozygotes, while C/C subjects in the obesity trial did not differ from T/T subjects in terms of weight loss or triglycerides. In an F2 intercross obtained by mating the OLETF and Fischer-344 (F344) rats, Ogino and co-workers [20] found that the Chr 3 QTL exhibits heterosis, heterozygotes showing significantly higher glucose levels than OLETF or F344 homozygotes. In our study, we detected two compound heteroses in the bovine PAPD1 gene that contribute to the extreme obesity-related phenotypes. The differences between PAPD1: c.10364AG/g.9367CG and PAPD1: c.10364GG/g.9367CG, and between PAPD1: c.10364AG/g.9419AC and PAPD1: c.10364GG/g.9419AC were 2.33 and 2.40 marbling scores, respectively (Figure 3A and 3B). In the Wagyu x Limousin F2 population, marbling scores ranged from 4 to 9.5 with a standard deviation of 1 marbling score. Therefore, these differences correspond to 2.33 and 2.40 standard deviations, indicating that double heterozygotes leads to extreme fat deposition, while individuals with other genotypes tend to be normal or slim. However, how the double heterosis produces high amount of intramuscular fat deposition need to be further examined.
Acknowledgements
We thank Dr. Michael MacNeil, USDA-ARS, Miles City, MT, for providing DNA and phenotypes for this research. Agricultural Research Center, Washington State University and Blue Rock Inc. supported this project.
Conflict of interests
The authors have declared that no conflict of interest exists.
References
1. National Task Force on the Prevention and Treatment of Obesity. Overweight, obesity, and health risk. Arch Intern Med. 2000 ;160(7):898-904
2. Spiegel AM, Alving BM. Executive summary of the Strategic Plan for National Institutes of Health Obesity Research. Am J Clin Nutr. 2005 ;82(1 Suppl):211S-214S
3. Perusse L, Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Snyder EE, Bouchard C. The human obesity gene map: the 2004 update. Obes Res. 2005 ;13(3):381-490
4. Flier JS. Obesity wars: molecular progress confronts an expanding epidemic. Cell. 2004 ;116(2):337-350
5. Mir PS, Mir Z, Kubert PS, Gaskins CT, Martin EL, Dodson MV, Calles JA, Johnson KA, Busboom JR, Wood AJ, Pittenger GJ, Reeves JJ. Growth, carcass characteristics, muscle conjugated linoleic acid (CLA) content, and response to intravenous glucose challenge in high percentage Wagyu, Wagyu x Limousin, and Limousin steers fed sunflower oil-containing diet. J Anim Sci. 2002 ;80(11):2996-3004
6. Hager J, Dina C, Francke S, Dubois S, Houari M, Vatin V, Vaillant E, Lorentz N, Basdevant A, Clement K, Guy-Grand B, Froguel P. A genome-wide scan for human obesity genes reveals a major susceptibility locus on chromosome 10. Nat Genet. 1998 ;20(3):304-308
7. Hinney A, Ziegler A, Oeffner F, Wedewardt C, Vogel M, Wulftange H, Geller F, Stubing K, Siegfried W, Goldschmidt HP, Remschmidt H, Hebebrand J. Independent confirmation of a major locus for obesity on chromosome 10. J Clin Endocrinol Metab. 2000 ;85(8):2962-2965
8. Price RA, Li WD, Bernstein A, Crystal A, Golding EM, Weisberg SJ, Zuckerman WA. A locus affecting obesity in human chromosome region 10p12. Diabetologia. 2001 ;44(3):363-366
9. Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 2004 ;32(20):6001-6014
10. Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J Biol Chem. 2005 ;280(20):19721-19727
11. Reichert AS, Neupert W. Mitochondriomics or what makes us breathe. Trends Genet. 2004 ;20(11):555-562
12. Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002 ;277(34):30409-30412
13. Wilson-Fritch L, Burkart A, Bell G, Mendelson K, Leszyk J, Nicoloro S, Czech M, Corvera S. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol. 2003 ;23(3):1085-1094
14. Jiang Z, Kunej T, Michal JJ, Gaskins CT, Reeves JJ, Busboom JR, Dovc P, Wright Jr RW. Significant associations of the mitochondrial transcription factor A promoter polymorphisms with marbling and subcutaneous fat depth in Wagyu x Limousin F2 crosses. Biochem Biophys Res Commun. 2005 ;334(2):516-523
15. Jiang Z, Kunej T, Wibowo TA, Michal JJ, Zhang ZW, Gaskins CT, Reeves JJ, Busboom JR, Dovc P, Wright Jr RW. The basal nucleus-encoded mitochondrial transcription genes and meat quality in beef cattle. Proceedings of 8th World Congress on Genetics Applied to Livestock Production. 2006 Belo Horizonte, Brazil
16. Saar K, Geller F, Ruschendorf F, Reis A, Friedel S, Schauble N, Nurnberg P, Siegfried W, Goldschmidt HP, Schafer H, Ziegler A, Remschmidt H, Hinney A, Hebebrand J. Genome scan for childhood and adolescent obesity in German families. Pediatrics. 2003 ;111(2):321-327
17. Boutin P, Dina C, Vasseur F, Dubois S, Corset L, Seron K, Bekris L, Cabellon J, Neve B, Vasseur-Delannoy V, Chikri M, Charles MA, Clement K, Lernmark A, Froguel P. GAD2 on chromosome 10p12 is a candidate gene for human obesity. PLoS Biol. 2003 ;1(3):E68
18. Vaisse C, Clement K, Durand E, Hercberg S, Guy-Grand B, Froguel P. Melanocortin-4 receptor mutations are a frequent and heterogeneous cause of morbid obesity. J Clin Invest. 2000 ;106(2):253-262
19. Pooley EC, Fairburn CG, Cooper Z, Sodhi MS, Cowen PJ, Harrison PJ. A 5-HT2C receptor promoter polymorphism (HTR2C - 759C/T) is associated with obesity in women, and with resistance to weight loss in heterozygotes. Am J Med Genet B Neuropsychiatr Genet. 2004 ;126(1):124-127
20. Ogino T, Moralejo DH, Zhu M, Toide K, Wei S, Wei K, Yamada T, Mizuno A, Matsumoto K, Shima K. Identification of possible quantitative trait loci responsible for hyperglycaemia after 70% pancreatectomy using a spontaneously diabetogenic rat. Genet Res. 1999 ;73(1):29-36
Author contact
![]() Correspondence to: Zhihua Jiang, Department of Animal Sciences, Washington State University, Pullman, WA 99164- 6351, USA. Tel: +509 335 8761; Fax: +509 335 4246; E-mail: jiangzedu
Correspondence to: Zhihua Jiang, Department of Animal Sciences, Washington State University, Pullman, WA 99164- 6351, USA. Tel: +509 335 8761; Fax: +509 335 4246; E-mail: jiangzedu
Received 2006-3-31
Accepted 2006-5-14
Published 2006-5-18