ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2008; 4(5):279-282. doi:10.7150/ijbs.4.279 This issue Cite
Short Research Communication
Discovery of four natural clones in a crayfish species Procambarus clarkii
1. Molecular population Genetics Group, Temasek Life Sciences Laboratory, 1 Research Link, National University of Singapore, 117604 Singapore
2. College of Animal Science and Technology, Nanjing Agriculture University, Nanjing, China
3. College of Animal Sciences, Zhejiang University, Hangzhou, China
Abstract
Self-cloning is quite rare in shrimp, lobsters, crayfish and crabs. Here we report the discovery of four natural clones of red swamp crayfish (Procambarus clarkii), each containing 2-6 genetically identical individuals, during the genotyping of 120 individuals with five microsatellites. The four clones were heterozygote at most of the five microsatellite loci. Phylogenetic analysis using microsatellite genotypes suggests recent origin of the four clones. Sequencing a part of the mitochondrial gene Cox I confirmed that the four clones were from the species Procambarus clarkii.
Keywords: crayfish, self-cloning, invasion, microsatellite
1. Introduction
Self-cloning, an asexual form of reproduction is found in some species of nematodes, gastropods, insects, fishes, amphibians and reptiles [1], while in crayfish species it was reported only in marbled crayfish in aquarium [2]. Red swamp crayfish, Procambarus clarkii native to south-central United States and Northeastern Mexico, has been transplanted to Hawaii, Japan, Europe and even the River Nile [3]. They were introduced to Nanjing, China from Japan in 1930s. This crayfish has rapidly spread to most provinces of China, and has established dense populations due to translocation by humans for food and lack of local predators [4]. Its production for human consumption reached 135, 000 tones in 2005. However, the species has caused serious problems to local fish, crustacean, aquatic plants and freshwater ecosystem, and their burrows lead to the loss of irrigation water and destroy rice field, leading to substantial economic loss in China [4, 5]. During the genotyping of 120 adult individuals collected in Nanjing, Jiangsu province and Tongxiang, Zhejiang province in China using five tetra-nucleotide microsatellites [6] to know the population structure of this alien invasive species, we surprisingly discovered four natural clones of P. clarkii. Here we report the discovery of the four clones in P. clarkii.
2. Materials and methods
Red swamp crayfish samples
We collected 62 and 58 adult P. clarkii individuals captured in ponds and ditches in two locations of China: Nanjing (Jiangsu province) and Tongxiang (Zhejing province). These individuals were sexed based on their external morphological genital opening located on the third pair of pereiopods in females, the fifth or last pair of pereiopods in males. A single third pleopod of every individual was collected and stored in absolute ethanol for DNA extraction [7].
Genotyping microsatellites
Five tetra-nucleotide microsatellites (PCLG03, PCLG07, PCLG09, PCLG12 and PCLG18) were selected from a published paper [6]. One primer of each pair was labeled with either FAM or Hex fluorescent dyes (1st Base). PCR amplification was performed on a PTC-100 machine (MJ Research) in 25 μl reaction volumes containing 40 ng DNA, 1 x PCR buffer including 1.5 mM MgCl2, 200 nM of each primer, 50 μM of each dNTP and one unit DNA polymerase (Finnzymes). Cycling conditions were as follows: 94°C for 2 min followed by 35 cycles of 94°C at 30 s, 50°C at 30 s and 72°C at 30 s, with a final extension at 72°C for 5 min. Sizes of the PCR products were determined against the size standard GS-ROX-500 (Applied Biosystems) using an ABI3730xl sequencer and its software, Genemapper v3.5 (Applied Biosystems) as described previously [7].
Sequencing part of the mitochondrial Cox 1 gene
The cytochrome c oxidase subunit 1 mitochondrial region (Cox I) is emerging as the standard barcode region for higher animals [8]. Therefore, the Cox I gene was used to confirm that the self-cloning crayfish was the red swamp crayfish in this study. To amplify the partial sequence of the mitochondrial Cox 1 gene, one pair of primers (F: TTTTTGGACACCCTGAAGTTTA, R: TATAAGCATCGGGATAATCTGAATA) were designed according to the known sequence (GenBank accession No. AY701195) of the gene of P. clarkii. PCR was conducted in 25 μl reaction volumes containing 40 ng total DNA, 200 nM of each primer, 200 μM dNTPs, one unit Taq DNA polymerase (Finnzymes) and 1x buffer with 1.5 mM MgCl2 on a PTC-100 PCR machine (MJ research). The PCR program was 94oC for 2 min, then 34 cycles of 94oC for 30 s, 55oC for 30 s and 72oC for 1 min, followed by a final extension of 72oC for 5 min. PCR products were cleaned using glassmilk (Bio 101) and sequenced in both 5' and 3' directions using the BigDye 3.0 chemicals (Applied Biosystems) and an ABI3730xl sequencer (Applied Biosystems). Forward and reverse sequences of each individual were assembled using Sequencher (GeneCodes).
Phylogenetic analysis
To confirm that the self-cloning crayfish is the red swamp crayfish, the partial sequences of the mitochondrial Cox I gene of 10 crayfish species (details see Figure 1) were downloaded from GenBank. The data set including the sequences of the four clones of P. clarkii collected in China was analyzed with maximum parsimony (MP), neighbor-joining (NJ), and minimum evolution (ME) as implemented in MEGA [9].
A minimum-evolution phylogenetic tree showing the relationships of crayfish species. The tree was constructed by a comparison of partial sequences of the mitochondrial Cox I gene. GenBank accession numbers, AY151515-AY151525 and DQ919058 for the self-cloning crayfish in China. Bootstrap values (> 66%) created by 1000 replicates are indicated on branches. Scale bar, 0.02 nucleotide substitutions/site. Parsimony and neighbor-joining analyses yielded the same topology. The phylogenetic analysis indicated the self-cloning crayfish was really the species Procambarus clarkii.

To understand the origin of the four clones, genetic relationships among the four clones and individuals sexually produced were analyzed based on microsatellite allele sharing index [10]. Phylogenetic (NJ and ME) trees were constructed as implemented in MEGA [9].
3. Results and discussion
Of the 62 individuals from Nanjing, 36 were females and 26 were males. From Tongxiang, 33 individuals were females while the remaining 25 were males. At the five microsatellite loci, a total of 49 alleles were detected. The Nanjing population showed more alleles than the Tongxiang population (49 vs. 35). The observed heterozygosity was 0.59 and 0.58 in the Nanjing and Tongxiang population, respectively, while the expected heterozygosity was 0.72 and 0.66 respectively. FST between the two populations was 0.06 (P < 0.05). We surprisingly found that four clones, containing five (clone 1), six (clone 2), two (clone 3) and two (clones 4) genetically identical female individuals, respectively (Table 1), while the remaining 105 individuals each showed a unique genotype group. Re-genotyping of DNA re-extracted from tissues by another person detected the same results, thus the possibility of DNA contamination would be excluded. According to the frequency of 49 alleles at the five loci in the 120 individuals, the possibility of two individuals sharing one genotype group was 1.47 x 10-5. Thus, these 15 individuals sharing the four genotype groups (clones) must be generated through self-cloning. These data indicate that self-cloning individuals of red swamp crayfish have already existed in freshwater ecosystems in China. Whether self-cloning individuals exist in other populations in China remains to investigate. The four clones were most probably diploid due to presence of one or two alleles per locus at all five loci (Table 1). The four clones showed higher observed heterozygosity (0.60-1.00) (Table 1) than the average observed heterozygosity (0.59) of individuals sexually produced in the two populations where the four clones came. Natural cloning could be generated by different forms of asexual reproduction including parthenogenesis [11], gynogenesis [12] and hybridogenesis [13]. However, hybridogenesis is hemiclonal, only half of the genome, the maternal one, is transmitted asexually. In the marbled crayfish [2], offspring were genetically identical to the parent, indicating it reproduces by apomixis [11], whereas, in Komodo dragons, offspring was instantaneous homozygosity of the entire genome produced by automixis with pure terminal fusion of nuclei [14]. Wolbachia infections have been reported to cause self-cloning in different invertebrate groups including arthropod species [15]. The self-cloning of P. clarkii may be generated by apomixis or automixis [16]. The high observed heterozygosity of the four clones in P. clarkii suggests that hybridization might also have played a major role in the formation of the four clonal lineages. However this speculation needs to be examined by further studies. The mechanisms underlying the self-cloning of P. clarkii remain to investigate.
Genotypes and heterozygosity of four natural clones of crayfish Procambarus clarkii at five microsatellite loci
| Clone | n | Microsatellite locus | Ho | ||||
|---|---|---|---|---|---|---|---|
| PCLG03 | PCLG07 | PCLG09 | PCLG13 | PCLG17 | |||
| 1 | 5 | 262/430 | 148/148 | 134/142 | 115/131 | 178/178 | 0.60 |
| 2 | 6 | 314/314 | 132/148 | 150/154 | 115/119 | 154/178 | 0.80 |
| 3 | 2 | 314/414 | 132/148 | 134/138 | 119/123 | 154/162 | 1.00 |
| 4 | 2 | 262/262 | 132/148 | 134/150 | 131/135 | 154/178 | 0.80 |
Clone 1 and 2 were detected from the samples collected in Tongxiang, Zhejiang province, while the clone 3 and 4 were from samples collected in Nanjing, China; n, the number of individuals sharing the genotype group; and Ho: observed heterozygosity.
To confirm that the four clones were from the species Procambarus clarkii, we sequenced the mitochondrial gene Cox I of all 15 self-cloning individuals and compared the sequences with those of other closely related crayfish species [2]. No nucleotide polymorphism in Cox 1 was detected in these 15 crayfish. Phylogenetic analysis [9] indicated that these self-cloning crayfish individuals were really from the species Procambarus clarkii (Figure 1). Phylogenetic analysis using microsatellite allele sharing index [10] revealed that the four clones were clustered with individuals sexually produced in different branches (Figure 2). No individual from the two locations shared any genotype group. These data suggest recent origin of the four clones. As self-cloning in this crayfish has not yet been observed in other countries, we suspected that self-cloning in this crayfish was induced by specific environment factors (e.g. Wolbachia infections) in China. The discovery of self-cloning in this crayfish species provides an opportunity to study environmental factors that induce self-cloning.
Self-cloning individuals could be more dangerous than crayfish sexually produced to freshwater ecosystem and rice field, as a single self-cloning individual may be enough to found a population. Hence, translocation of this crayfish among different regions must be banned to prevent its spread, and self-cloning individuals in the wild must be identified and eradicated in situ.
Genetic relationships among the four natural clones and 16 individuals sexually produced of red swamp crayfish as revealed by the proportion of shared alleles at five microsatellite loci. TX, individuals collected from Tongxiang, Zhejiang province; NJ, individuals collected from Nanjing, China. Scale bar representing genetic dissimilarity is shown under the tree. The four clones were clustered with individuals sexually produced in different branches, and no individuals from the two locations shared any genotype group, suggesting recent origin of the four clones.
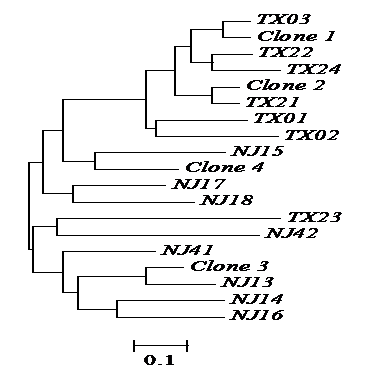
Acknowledgements
We thank G. Lin, F. Feng and J. Li for technical assistance. This study was supported by the internal research funds from the Temasek Life Sciences Laboratory, Singapore.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Dawley RM, Bogart JP. Evolution and Ecology of Unisexual Vertebrates. Albany, New York: New York State Museum. 1989
2. Scholtz G. et al. Parthenogenesis in an outsider crayfish. Nature. 2003;421:806
3. Huner JV. Procambarus in North America and elsewhere. In: (ed.) Holdich DM, Lowery RS. Freshwater Crayfish: Biology, Management and Exploitation. Timber Press: Portland. 1988:239-261
4. Li SC. et al. Investigation on and analysis of alien invasions in Chinese farming industry. Chin Agri Sci Bul. 2005;21:156-159
5. Li ZY, Xie Y. Invasive Alien Species in China Beijing. Forestry Press. 2002
6. Belfiore NM, May B. Variable microsatellite loci in red swamp crayfish, Procambarus clarkii, and their characterization in other crayfish taxa. Mol Ecol. 2000;9:2231-2234
7. Zhu ZY. et al. Isolation and characterization of polymorphic microsatellites from red coral grouper (Plectropomus maculatus). Mol Ecol Notes. 2005;5:579-581
8. Meusnier I. et al. A universal DNA mini-barcode for biodiversity analysis. BMC genomics. 2008;9:214
9. Kumar S, Tamura K, Nei M. Integrated software for molecular evolutionary genetics analysis and sequence alignment. Bioinformatics. 2004;5:150-163
10. Chakraborty R, Jin L. Determination of relatedness between individuals using DNA fingerprinting. Hum Biol. 1993;65:875-895
11. Martin P, Kohlmann K, Scholtz G. The parthenogenetic Marmorkrebs (marbled crayfish) produces genetically uniform offspring. Naturwissenschaften. 2007;94:843-846
12. Beukeboom LW, Vrijenhoek RC. Evolutionary genetics and ecology of sperm-dependent parthenogenesis. J Evol Biol. 1998;11:755-782
13. Marracci S, Ragghianti M. The hybridogenetic Rana (Pelophylax) esculenta complex studied in a molecular context. Italian J Zool. 2008;75:109-112
14. Watts PC. et al. Parthenogenesis in Komodo dragons. Nature. 2006;444:1021-1022
15. Bourtzis K, O'Neill S. Wolbachia infections and arthropod reproduction. Bioscience. 1998;48:287-293
16. Judsona OP, Normark BB. Ancient asexual scandals. Trend Ecol Evol. 1996;11:41-46
Author contact
![]() Correspondence to: Dr Gen Hua Yue; Tel: +65-68727405; Fax: +65-68727007; Email: genhuaorg.sg
Correspondence to: Dr Gen Hua Yue; Tel: +65-68727405; Fax: +65-68727007; Email: genhuaorg.sg
Received 2008-6-27
Accepted 2008-8-26
Published 2008-8-28
