ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2023; 19(14):4627-4643. doi:10.7150/ijbs.85236 This issue Cite
Research Paper
Activation of STING by SAMHD1 Deficiency Promotes PANoptosis and Enhances Efficacy of PD-L1 Blockade in Diffuse Large B-cell Lymphoma
1. Department of Hematology, Shandong Provincial Hospital, Shandong University, Jinan, Shandong, 250021, China.
2. Department of Hematology, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, Shandong, 250021, China.
3. Shandong Provincial Engineering Research Center of Lymphoma, Jinan, Shandong, 250021, China.
4. Branch of National Clinical Research Center for Hematologic Diseases, Jinan, Shandong, 250021, China.
5. National Clinical Research Center for Hematologic Diseases, the First Affiliated Hospital of Soochow University, Suzhou, 251006, China.
Abstract
Genomic instability is a significant driver of cancer. As the sensor of cytosolic DNA, the cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) pathway plays a critical role in regulating anti-tumor immunity and cell death. However, the role and regulatory mechanisms of STING in diffuse large B-cell lymphoma (DLBCL) are still undefined. In this study, we reported that sterile alpha motif and HD domain-containing protein 1 (SAMHD1) deficiency induced STING expression and inhibited tumor growth in DLBCL. High level of SAMHD1 was associated with poor prognosis in DLBCL patients. Down-regulation of SAMHD1 inhibited DLBCL cell proliferation both in vitro and in vivo. Moreover, we found that SAMHD1 deficiency induced DNA damage and promoted the expression of DNA damage adaptor STING. STING overexpression promoted the formation of Caspase 8/RIPK3/ASC, further leading to MLKL phosphorylation, Caspase 3 cleavage, and GSDME cleavage. Up-regulation of necroptotic, apoptotic, and pyroptotic effectors indicated STING-mediated PANoptosis. Finally, we demonstrated that the STING agonist, DMXAA, enhanced the efficacy of a PD-L1 inhibitor in DLBCL. Our findings highlight the important role of STING-mediated PANoptosis in restricting DLBCL progression and provide a potential strategy for enhancing the efficacy of immune checkpoint inhibitor agents in DLBCL.
Keywords: DLBCL, SAMHD1, STING, PANoptosis, DMXAA.
Introduction
As the most common subtype of B-cell lymphoma, diffuse large B-cell lymphoma (DLBCL) is featured by differential clinical, immunophenotypic, cytogenetic, and genetic characteristics [1]. Despite the recent improvement with new treatments [2], 30%-40% of patients are insensitive to standard chemoimmunotherapy (CIT) and eventually progress to relapsed/refractory (R/R) stages [3]. Patients that are resistant to CIT may benefit from other treatment options, including antibody-drug conjugates (ADC), immune checkpoint inhibitors, and chimeric antigen receptor T (CAR-T) cell therapy, all of which have been developed and incorporated into clinical practice; however, variable treatment responses are observed [4, 5]. Therefore, new treatments are still urgently needed. Understanding underlying pathogenesis and identifying new biology and targets are critical for any new therapy discovery and development. Given that the malignant transformation of B-cells is correlated with the extensive DNA editing in the germinal center, targeting DNA damage response (DDR) can potentially be an important strategy for the development of new treatment for DLBCL [6, 7].
As a deoxynucleoside triphosphate hydrolase (dNTPase), sterile alpha motif and HD domain-containing protein 1 (SAMHD1) plays an important role in genomic stability by balancing the DNA precursor pools [8]. In particular, SAMHD1 mainly functions by promoting the end resection process in DNA repair [9] and resolving stalled replication forks in DNA replication [10]. As such, dysregulation of SAMHD1 is linked to cell cycle regulation and chemosensitivity to nucleoside analogs in human cancers [11, 12]. A recent study demonstrated that targeted inhibition of SAMHD1 by hydroxyurea was a safe and effective strategy in a phase 1 trial [13], supporting an important role for SAMHD1 in hematological tumors.
It is well established that the cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) signaling pathway is critical for DNA damage perceiving [14]. Induction of STING-associated inflammation facilitates the interferon (IFN)-related innate immunity priming, which is identified as a barrier to tumorigenesis [15]. Interestingly, targeting DDR may induce anti-tumor immune responses via activating the cGAS-STING axis, and further enhance the efficacy of immunotherapy [16, 17]. Hence, understanding the interplay between the DDR and the cGAS-STING pathway may provide a targeting strategy for the development of effective treatment for lymphoma patients [18].
In addition to immune regulation, the cGAS-STING pathway mediates cell death, including apoptosis, autophagy, necroptosis, pyroptosis, and ferroptosis [19, 20]. Necroptosis and pyroptosis are the newly discovered non-apoptotic forms of cell death, characterized by the loss of cell membrane integrity and the release of cytoplasmic content [21]. Necroptosis depends on the activation of mixed lineage kinase domain-like protein (MLKL), while pyroptosis relies on the cleavage of gasdermin proteins. In particular, interactions between necroptosis, apoptosis, and pyroptosis were referred to PANoptosis [22, 23]. As a programmed cell death (PCD), PANoptosis depends on the PANoptosome, which contains multiple proteins essential for activating various cell death effectors [24, 25]. Increasing evidence shows that PANoptosis is deeply related to tumorigenesis and immune responses [26, 27]. Therefore, PANoptosis induction is considered a viable strategy in reducing tumor burden and resolving treatment resistance [28, 29]. Given the importance of PANoptosis, we attempt to explore the mechanism of PANoptosis in DLBCL and hope to identify a trackable target for therapeutic interventions.
Here, we investigate the role of STING in DLBCL tumor growth and anti-DLBCL treatment. STING is activated in SAMHD1-deficient DLBCL cells, and further induces PANoptosis to suppress cell growth. Specifically, STING-mediated PANoptosis depends on the activation of MLKL, Caspase 3 (CASP3), and gasdermin E (GSDME). Moreover, we demonstrate that STING agonist DMXAA enhances programmed death ligand 1 (PD-L1) blockade efficacy. Our results highlight that targeted activation of STING holds promise in treating DLBCL.
Materials and Methods
Lymph node samples and peripheral blood samples
Lymph node samples were collected from 100 newly diagnosed DLBCL patients and 20 lymphoid reactive hyperplasia (RHL) patients. The diagnostic criteria were established according to the World Health Organization (WHO) classification [30]. Among them, 65 enrolled DLBCL patients received treatment, of which 49 documented the treatment regimens and response (Table S1). Besides, the response was evaluated after every three courses of treatment, mainly based on positron emission tomography/computed tomography (PET/CT) results (Table S1). Peripheral blood mononuclear cells (PBMCs) of healthy donors were isolated by the Ficoll-Hypaque density gradient centrifugation method (TBD Science, Tianjin, China) and naïve CD19+ B-cells were separated by CD19+ magnetic microbeads kit (Miltenyi Biotec, Bergisch Gladbach, Germany). The purity of enriched CD19+ B-cells was detected by flow cytometry (FCM) analysis, as previously reported (Figure S1A) [31].
CRISPR/CRISPR associated 9 (Cas9)-generated STING-knockout (KO) cells
CRISPR/Cas9 genomic editing system was applied to generate STING-KO cells. Stably expressing Cas9-gRNA was established and packaged by OBiO Technology Corp., Ltd. (Shanghai, China). The viral vector of gRNA for STING deletion was as follows: pLenti-U6-spgRNA (TMEM173)-CMV-Puro-P2A-3xFLAG-spCas9 WPRE. Vector control was annotated as WT. After 72 hours of transfection, cells were treated with 2.0μg/ml puromycin for 3 days, and single cell was screened by a limited dilution method.
Lentiviral generation and cell transfection
Lentivirus vectors that encoded SAMHD1-knockdown (KD), SAMHD1-overexpression (LV-SAMHD1), STING-overexpression (LV-STING), and empty vectors (Ctrl and LV-Con) were constructed by GeneChem (Shanghai, China). Lentivirus infection was carried out according to the manufacturer's instruction at a multiplicity of infection (MOI) =100. Transfected cells were selected with puromycin (2.0 μg/ml) after 72 hours of transfection. Transfection efficiency was then verified by immunoblot. SAMHD1-KD, Ctrl and LV-Con sequences were available in Table S2.
RNA-sequencing (RNA-seq) and bioinformatics analysis
For RNA-seq, total RNA was isolated with TRIzol reagent (Invitrogen, 15596026, CA, USA) from SAMHD1-KD LY1 cells. Novogene (Beijing, China) performed the RNA-seq experiments. Briefly, sequencing libraries were generated from purified mRNA, after which the library preparations were sequenced on an Illumina HiSeq platform and generated 150 bp paired-end reads. HTSeq v0.6.0 was then applied to calculate the numbers of reads and the fragments per kilobase million (FPKM). Gene ontology (GO), Reactome, gene set enrichment analysis (GSEA), and differential expression analyses were finally performed by R language. In addition, SAMHD1 expression in DLBCL samples was derived from the Oncomine and Cancer Genome Atlas (TCGA) databases.
Immunofluorescence (IF)
Ctrl and SAMHD1-KD cells were mounted on glass slides, fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and then blocked with 10% goat serum in PBS. After incubating with primary antibodies, cells were probed with fluorescent secondary antibodies and stained by DAPI (Solarbio, S2110, Beijing, China). Confocal microscopic images were captured by Leica TCS SP8 MP (Leica, Wetzlar, Germany) and measured by the Image J software (National Institutes of Health, USA). The antibodies applied in IF were listed in Table S3.
Neutral comet assay
Neutral comet assay was performed on Ctrl and SAMHD1-KD cells according to the manufacturer's protocol (Trevigen, Maryland, USA). DAPI (Solarbio, S2110) was used to stain the DNA fragments. Images were collected by Olympus BX51 fluorescence microscopy with ×200 objectives. Tail moments were analyzed by Open Comet software (Cambridge, USA). Fifty individual cells were counted in each experiment.
Co-immunoprecipitation (CO-IP)
LY1 and LY3 cells transfected with LV-STING were lysed with the RIPA buffer (Beyotime, P0013, Shanghai, China). Protein lysates incubated with 3 μg of the anti-Caspase 8 (CASP8)-mouse antibody (Proteintech, 66093-1-Ig, IL, USA) or normal mouse IgG antibody (Santa Cruz, sc-2025, CA, USA) were rotated overnight at 4℃. The immune complexes were then treated with 20μl protein A/G PLUS-agarose beads (Santa Cruz, sc-2003) for 4 hours at 4℃. Beads bound to immunoprecipitates were denatured by a mental bath, followed by immunoblot analysis.
Lactate dehydrogenase (LDH) release assay and morphology observation
According to the manufacturer's instructions, a CytoTox 96 non-radioactive cytotoxicity assay kit (Promega, G1780, Madison, USA) was performed to test supernatant LDH levels of cells. For morphological investigation, DLBCL cells transfected with lentivirus vectors were visualized by the OLYMPUS CKX41 inverted microscope at ×200 objectives in three different fields. Image processing was performed by Image J software.
Immunohistochemistry (IHC)
IHC staining was performed as previously introduced [31]. The number of positive cells referred to the expression level. The receiver operating characteristic (ROC) curve was constructed by the numbers of positive cells, of which the Youden's index was recognized as the evaluation standard (value = 0.337). With these criteria, tissues were interpreted as “positive (positive rate > 33.7%)” or “negative (positive rate ≤ 33.7%).” The antibody applied in IHC was Rabbit anti-SAMHD1 (1:200, Proteintech, 12586-1-AP).
Cell lines and reagents
Human DLBCL cell lines LY1, LY3, and LY8 were cultured in Iscove's modified Dulbecco's medium (IMDM) (Gibco, CA, USA) supplemented with 10% heat-inactivated fetal bovine serum (HyClone, UT, USA). Cells were left to incubate at 37°C in a humidified 5% CO2 atmosphere. DMXAA (S1537) and BMS1166 (S8859) were obtained from Selleckchem (TX, USA). CalcuSyn software (Cambridge, UK) was applied to evaluate the effectiveness of drug combinations, representing by the combination index (CI) values. In particular, CI < 1 indicates synergism, CI=1 shows an additive effect, and CI > 1 represents antagonism.
Cell viability detection
Cell viability was measured by the cell counting kit-8 (CCK-8) assay (Dojindo, CK04, MD, USA). In briefly, cells were seeded in 96-well plates at 1×104 cells/well and stained with 10μl CCK-8/well at a certain point in time, after which optical density was detected at 450nm by Multiskan GO Microplate Reader (Thermo Scientific, Rockford, IL, USA).
Western blotting (WB)
WB was performed as previously introduced [32]. Cell lysates were obtained using the RIPA buffer (Beyotime, P0013B) mixed with phosphatase inhibitor cocktail (PhosSTOP, Roche, Basel, Switzerland). Total proteins were loaded in the SDS-PAGE (Bio‐Rad, California, USA) and transferred to polyvinylidene fluoride (PVDF) membranes. After blocking with 10% skimmed milk, PVDF membranes were incubated with primary and secondary antibodies. The antibodies were listed in Table S4.
FCM
Apoptosis rates were detected by Annexin V-PE/7AAD or Annexin V-FITC/PI double-staining FCM (BD Biosciences, Bedford, MA, USA). The percentage of apoptotic cells was conducted on Navios Flow Cytometer (Beckman Coulter, CA, USA).
In vivo mice xenograft models
Animal experimental procedures were performed in accordance with protocols approved by the Institutional Animal Care and Research Advisory Committee of Shandong Provincial Hospital. Mice were purchased from the Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). For the in vivo study of SAMHD1, a total of 1×107 Ctrl or SAMHD1-KD LY1 cells were subcutaneously injected into the right upper flanks of severe combined immunodeficiency (SCID) beige mice (n = 6/group), respectively. For the efficacy of drug combination studies, xenograft models were established in 4-week-old female Balb/c nude mice by injecting 1×107 wild-type LY1 cells into the subcutaneous of right lower flanks. When tumors reached 100 mm3, tumor-bearing mice were randomized into specified groups (n = 6/group). For monotherapy, the mice were intra-peritoneally injected with 100μl BMS1166 (250 μg/ml) or DMXAA (20 mg/kg). For combination therapy, BMS1166 (250 μg/ml) and DMXAA (20 mg/kg) were alternately injected every other day. Tumor dimensions were measured every 2 days, which were calculated using the equation V = (l × w2) × 0.5. Mice were sacrificed until any one of several criteria were met, including severe lethargy, more than 10% body-weight loss, and approximately 2cm of tumor diameter.
Statistical analysis
Data was presented as mean ± standard deviation (SD) from at least three separate experiments. All statistical analyses were performed with SPSS Statistics version 20.0 and the GraphPad Prism software (version 8.0a, La Jolla, CA, USA). Statistical significance between the two groups was determined by unpaired two-tailed t-test with assumed normal distribution. If normality tests failed, Mann-Whitney tests were applied. Three or more groups were analyzed by Welch's one-way ANOVA analysis with Dunnett's T3 tests. Proliferation curves were measured by two-way ANOVA analysis with Sidak correction. Log-rank tests were used in survival analysis. Contingency tables were determined by two-tailed χ2 test. P-value < 0.05 was considered as statistical significance, including *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Results
SAMHD1 is highly expressed in DLBCL and associated with poor prognosis of patients
We first sought to determine the expression of SAMHD1 in DLBCL patients. Analyses of the Oncomine and TCGA databases revealed a significant increase of SAMHD1 mRNA in DLBCL tissues, compared with normal B-cell subtypes (Figure 1A, B). Subsequently, the protein levels of SAMHD1 in DLBCL tissues were determined by IHC in 100 DLBCL patients. Specifically, the positive expression of SAMHD1 was significantly higher in DLBCL patients (positive rate = 60.0%, 60/100) than that in RHL tissues (positive rate = 35.0%, 7/20) (Figure 1C). In addition, SAMHD1 expression showed no difference in DLBCL samples with different cell-of-origin (COO) (Figure 1C).
To further investigate the clinical significance of SAMHD1 expression in DLBCL patients, we analyzed the clinical information of patients enrolled in IHC staining (Table 1). Interestingly, SAMHD1-positive patients were featured with defective B symptoms and decreased serum LDH (Table 1). Although SAMHD1 was associated with antiviral infection, SAMHD1 expression showed no correlation with Epstein-Barr virus (EBV) or Hepatitis B virus (HBV) infections in DLBCL patients (Table 1). Analysis of clinical treatment responses was also performed to identify the effects of SAMHD1 on therapeutic efficacy. Forty-nine enrolled patients received systematic treatment and effect evaluation, and 71.4% of them (10/14) achieved complete remission (CR) after treatment with six cycles of rituximab, cyclophosphamide, doxorubicin, vincristine, and oral prednisone (R-CHOP) (Table S1). However, differential expression of SAMHD1 showed no significant impact on clinical efficacy (Table 1). Subsequent survival analysis revealed a poorer prognosis in SAMHD1-positive DLBCL patients (Figure 1D), indicating that high SAMHD1 expression led to inferior prognosis in DLBCL patients.
SAMHD1 expression is up-regulated in DLBCL and related to tumor growth. A, B. The mRNA levels of SAMHD1 in DLBCL samples from the Oncomine (A) and TCGA database (B). C. IHC staining revealed the expression of SAMHD1 in GCB-like DLBCL, ABC-like DLBCL, and RHL tissues (upper), followed by relative quantitative analysis (lower, p=0.04). P value came from the Chi-square test. D. Kaplan Meier plots for DLBCL patients enrolled in IHC staining (n=100, p=0.03). E. Protein levels of SAMHD1 in normal CD19+ B-cells (N1, N2, N3) and DLBCL cell lines (LY1, LY3, LY8) were detected by western blotting. F, G. LY1 cells were transfected with LV-Con or LV-SAMHD1 sequences. After stable transfection, transfection efficiency was examined by immunoblot (F), while cell viability was detected by CCK-8 assay (G). H, I. LY1 and LY3 cells were transfected with Ctrl or SAMHD1-KD sequences (sh1#, and sh2#). After stable transfection, knockdown efficiency was examined by immunoblot (H), while cell viability was detected by CCK-8 assay (I). J-L. Schematic of in vivo tumor growth investigation (J). SCID beige mice were injected with Ctrl or SAMHD1-KD LY1 cells (n=6/group). Tumor bodies were taken on day 28 (K). Tumor growth curves were shown from day 18 to day 28 (L). Immunoblot images in E, F, and H were the representation of 3 independent experiments. Vertical bars indicated mean ± SD. P values came from Kruskal-Wallis test followed by Dunn's test (A), Log-rank test (D), unpaired two-tailed t-test (B, F, G, H, I), and Two-way ANOVA with Sidak correction (L). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.

Correlation between SAMHD1 protein expression and clinicopathologic parameters of DLBCL patients.
| Variables | Number of Patients | SAMHD1 expression | p value | |
|---|---|---|---|---|
| Positive | Negative | |||
| Age (years) | ||||
| ≤ 60 | 50 | 30 (60.0%) | 20 (40.0%) | 1.000 |
| > 60 | 50 | 30 (60.0%) | 20 (40.0%) | |
| Gender | ||||
| Male | 52 | 31 (59.6%) | 21 (40.4%) | 0.935 |
| Female | 48 | 29 (60.4%) | 19 (39.6%) | |
| Subtype | ||||
| GCB | 40 | 20 (50.0%) | 20 (50.0%) | 0.248 |
| ABC | 60 | 37 (61.7%) | 23 (38.3%) | |
| Ann Arbor stage | ||||
| I or II | 37 | 26 (70.3%) | 11 (29.7%) | 0.108 |
| III or IV | 63 | 34 (54.0%) | 29 (46.0%) | |
| B symptoms | ||||
| Present | 19 | 7 (36.8%) | 12 (63.2%) | 0.022* |
| Absent | 81 | 53 (65.4%) | 28 (34.6%) | |
| Serum LDH levels | ||||
| Normal (LDH-ratio ≤ 1) | 53 | 38 (71.7%) | 15 (28.3%) | 0.011* |
| Elevation (LDH-ratio > 1) | 47 | 22 (46.8%) | 25 (53.2%) | |
| ENI | ||||
| Involved site (≤ 1) | 24 | 16 (66.7%) | 8 (33.3%) | 0.131 |
| Involved site (> 1) | 50 | 24 (48.0%) | 26 (52.0%) | |
| IPI Score | ||||
| 0-2 | 50 | 33 (66.0%) | 17 (34.0%) | 0.221 |
| 3-5 | 50 | 27 (54.0%) | 23 (46.0%) | |
| EBV infection | ||||
| Positive | 9 | 5 (55.6%) | 4 (44.4%) | 0.821 |
| Negative | 37 | 19 (51.4%) | 18 (48.6%) | |
| HBV infection | ||||
| Positive | 27 | 17 (63.0%) | 10 (37.0%) | 0.684 |
| Negative | 60 | 35 (58.3%) | 25 (41.7%) | |
| Treatment response after six cycles of R-CHOP | ||||
| CR and PR | 9 | 8 (88.9%) | 1 (11.1%) | 0.505 |
| PD | 5 | 3 (60.0%) | 2 (40.0%) | |
*GCB: Germinal center B-cell; ABC: Activated B-cell; LDH: Lactate dehydrogenase; IPI: International Prognostic Index. B symptoms refer to the fever of unknown cause >38℃ for three consecutive days, night sweats, or weight loss >10% within six months. Extranodal involvement (ENI) excludes testicle, central nervous system and bone marrow due to the significant impact of survival. EBV: Epstein-Barr virus. HBV: Hepatitis B virus. R-CHOP: Rituximab, cyclophosphamide, doxorubicin, vincristine, oral prednisone. CR: Complete remission. PR: Partial remission. PD: Progressive disease
SAMHD1 expression is important for DLBCL cell growth
After establishing the correlation between the SAMHD1 expression and clinical outcomes of DLBCL patients, we attempted to explore the role of SAMHD1 in tumorigenesis. To select proper cell models, immunoblot analysis was used to determine the protein levels of SAMHD1 in DLBCL cell lines. As shown in Figure 1E, SAMHD1 expression was increased in several DLBCL cell lines, especially in LY1 and LY3 cells. To verify whether SAMHD1 expression contributed to cell survival, we constructed SAMHD1-overexpressed LY1 cells utilizing the LV-SAMHD1 sequence (Figure 1F). As expected, cell viability was increased with SAMHD1 overexpression (Figure 1G). To confirm that these findings were due to SAMHD1 expression, SAMHD1-KD models were established in LY1 and LY3 cells. Individual shRNA sequence sh2# with significant knockdown effects was selected for the functional analysis (Figure 1H). In contrast to SAMHD1 overexpression, SAMHD1 deficiency significantly suppressed cell viability (Figure 1I).
To further investigate the role of SAMHD1 in vivo, we examined the tumor growth in a xenograft mouse model established by Ctrl and SAMHD1-KD LY1 cells (Figure 1J). SAMHD1 deficiency markedly reduced tumor size and suppressed tumor growth (Figure 1K, L).
SAMHD1 deficiency induces DNA damage and double-strand DNA (dsDNA) accumulation
Next, we sought to explore the mechanism by which SAMHD1 deficiency suppressed DLBCL tumor growth. Our analysis with FCM detected a significant increase in apoptosis rates in SAMHD1-KD DLBCL cells (Figure S2A). Additionally, optical microscopy revealed marked morphological changes in these cells, including cell shrinkage and cell membrane swelling (Figure S2B). Morphological changes suggested the impairment of cell membrane integrity in SAMHD1-KD DLBCL cells, which was confirmed by the high levels of supernatant LDH (Figure S2C). Given the specific changes in phenotypes, we hypothesized that down-regulation of SAMHD1 in DLBCL cells led to cell death.
To further explore the underlying mechanisms of SAMHD1-mediated cell death, we performed RNA-seq in SAMHD1-deficient LY1 cells and screened the differentially expressed genes (DEGs) (Figure 2A). GO analysis displayed that DEGs were enriched in the chromosomal regions, nuclear chromatin, replication forks, DNA damage, and double-strand break (DSB) sites (Figure 2B). Reactome analysis further identified the enrichment in biochemical reactions related to DNA damage repair, mainly including DSB repair, homologous recombination repair, nucleotide metabolism, and cell cycle checkpoints (Figure 2C).
To verify the generation of DNA damage, we investigated the density of DNA damage in SAMHD1-deficient DLBCL cells. H2AX and Hsp60, the DNA damage markers, were used to locate damage sites. Of note, down-regulation of SAMHD1 dramatically increased the intensity of p-H2AX in the nucleus (Figure 2D); however, the intensity of cytosolic Hsp60 showed no significant changes (Figure S3A). Along with DNA damage, neutral comet assay detected the extended tails in SAMHD1-deficent cells (Figure 2E), suggesting the accumulation of DNA fragments. Besides, cytosolic dsDNA was increased in these cells (Figure 2F). Thus, SAMHD1 deficiency leads to DNA damage and dsDNA accumulation in DLBCL cells.
SAMHD1 deficiency induces DNA damage and dsDNA accumulation. A-C. RNA-seq was performed in Ctrl and SAMHD1-KD LY1 cells (three biological replicates for each group). Volcano plot revealed DEGs in SAMHD1-KD cells compared with Ctrl cells (A). Bubble plots revealed the significantly up-regulated GO terms (B) and Reactome pathways (C) in SAMHD1-KD LY1 cells. D. Fluorescence plots (upper) and fluorescence density (lower) of H2AX in Ctrl and SAMHD1-KD DLBCL cells. Scale bar=10μm. E. Representative images (upper) and qualification (lower) of neutral comet assay revealed the abundance of DNA fragments in Ctrl and SAMHD1-KD DLBCL cells. Scale bar=20μm. Individual dots represented single cells. F. Fluorescence plots (upper) and fluorescence density (lower) of dsDNA in Ctrl and SAMHD1-KD DLBCL cells. Scale bar=10μm. Vertical bars indicated mean ± SD. P values from unpaired two-tailed t-test (D, E, F). *p<0.05, ****p<0.0001.

Down-regulation of SAMHD1 promotes the expression of the STING-related DNA-sensing pathway. A. GSEA plot of the cytosolic DNA-sensing pathway (NES=1.36, p<0.05, FDR=0.07). B. Immunoblot showed the protein levels of cGAS and STING proteins in Ctrl and SAMHD1-KD DLBCL cells. Immunoblot images were the representation of 3 independent experiments. C. Heatmap revealed the DEGs in the cytosolic DNA-sensing pathway between Ctrl and SAMHD1-KD LY1 cells.

Down-regulation of SAMHD1 promotes the expression of the STING-related DNA-sensing pathway
As SAMHD1 deficiency induced DNA damage, we further examined the expression of DNA-sensing pathways in SAMHD1-deficient cells. GSEA analysis revealed the up-regulation of the STING-mediated cytosolic DNA-sensing pathway in SAMHD1-KD LY1 cells (p<0.05; Figure 3A). However, immunoblot analysis verified that SAMHD1 deficiency mainly promoted the expression of STING, rather than cGAS (Figure 3B). To explore the biological functions of SAMHD1-KD-induced STING, we analyzed the DEGs in the cytosolic DNA-sensing pathway. Although STING was involved in innate immune activation [33], IFN regulators were inactivated in SAMHD1-KD DLBCL cells, including TANK-binding kinase 1 (TBK1) and interferon regulatory factor 3 (IRF3) (Figure 3C, Figure S4A). Interestingly, SAMHD1 deficiency promoted the transcription of genes related to cell death, including receptor-interacting protein kinase 1 (RIPK1), receptor-interacting protein kinase 3 (RIPK3), nuclear factor kappa-B (NF-κB), and apoptosis-associated speck-like protein containing CARD (ASC) (Log2FoldChange > 2, p < 0.05; Figure 3C). These data suggest the correlation between STING and cell death regulation.
STING activation induces multiple forms of cell death to suppress DLBCL cell growth
To investigate whether cell death was mediated by STING, we constructed lentivirus-mediated LV-STING and CRISPR/Cas9-mediated STING-KO models in LY1 and LY3 cells, respectively (Figure 4A, B). As shown in the FCM analysis, STING overexpression significantly increased the percentage of Annexin V+/7AAD- and Annexin V+/7AAD+ cells (Figure 4C). In contrast, Annexin V-FITC+/PI- and Annexin V-FITC+/PI+ cells were decreased by STING deletion (Figure 4D). These results were indicative of STING activation-induced cell apoptosis. Subsequently, morphological changes and LDH release were detected to verify the generation of non-apoptotic cell death. It was worth noting that LV-STING cells, rather than STING-KO cells, were featured with cell membrane swelling and high levels of supernatant LDH (Figure 4E, F).
STING activation induces multiple forms of cell death to suppress DLBCL cell growth. A. STING overexpression model was established in LY1 and LY3 cells utilizing LV-STING sequences. Transfection efficiency was determined by immunoblot. B. STING-KO model was constructed in LY1 and LY3 cells by transfecting three independent CRISPR/Cas9-mediated STING-KO sequences (STING-KO #1, #2, and #3). Transfection efficiency was determined by immunoblot. C. Annexin V-PE/7AAD double staining flow cytometry revealed the scatter plots (left) and quantitative apoptosis rates (right) in LV-Con and LV-STING DLBCL cells. D. Annexin V-FITC/PI double staining flow cytometry revealed the scatter plots (left) and quantitative apoptosis rates (right) in WT and STING-KO DLBCL cells. E. Microscopic images (left, scale bar=50μm) and supernatant LDH levels (right) of LV-Con and LV-STING DLBCL cells. White arrows indicated cell membrane swelling. F. Microscopic images (left, scale bar=50μm) and supernatant LDH levels (right) of WT and STING-KO DLBCL cells. G. CCK-8 assay revealed the cell viability of LV-STING (left) and STING-KO (right) DLBCL cells, which were compared with empty vectors. H, I. Ctrl and SAMHD1-KD sequences were transfected in STING-KO DLBCL cells. Transfection efficiency was determined by immunoblot (H). Cell proliferation of Ctrl, SAMHD1-KD, Ctrl+STING-KO, and SAMHD1-KD+STING-KO DLBCL cells was compared by CCK-8 assay (I). Immunoblot images in A, B, and H were the representation of 3 independent experiments. Vertical bars indicated mean ± SD. P values from unpaired two-tailed t-test (A, C, D, E, F, and G) and Two-way ANOVA with Sidak correction (I). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, and ns=no significance.

Given the functions of STING in cell death induction, we further explored whether STING overexpression suppressed DLBCL cell growth. As expected, STING overexpression decreased cell viability, while STING deletion enhanced cell viability (Figure 4G). To better understand whether STING was involved in SAMHD1-mediated cell survival, intrinsic STING was deleted in DLBCL cells with and without SAMHD1-KD (Figure 4H). Genetic deletion of STING in SAMHD1-KD cells restored cell survival to a similar level of Ctrl cells, suggesting that STING mediated SAMHD1 deficiency-induced cell death (Figure 4I). Together we demonstrate the functions of STING in DLBCL, that is, up-regulation of STING suppresses DLBCL cell growth by inducing various forms of cell death.
STING activates MLKL, CASP3, and GSDME to induce PANoptosis
Next, we attempted to explore the underlying mechanisms of STING-induced cell death. STING is critical in driving PCD, including necroptosis, apoptosis, and pyroptosis [34]. To determine the modes of STING-induced cell death, we performed immunoblot analysis to verify the downstream pathways in LV-STING DLBCL cells. RNA-seq analyses previously revealed an increase in RIPK1 and RIPK3, the major regulators of necroptosis [35, 36]. Consistently, phosphorylated RIPK3 and MLKL were increased in DLBCL cells with overexpressed STING, indicative of necroptosis induction (Figure 5A). In addition to necroptosis, we also detected caspase-dependent apoptosis and pyroptosis. Of note, STING overexpression promoted the activation of apoptotic effectors, represented by the cleavage of CASP8 and CASP3 (Figure 5B). Pyroptosis is a non-apoptotic cell death that relies on caspase-mediated gasdermin cleavage. As the specific gasdermin protein in cancer cells, GSDME responds to CASP3 and further induces secondary pyroptotic cell death [37]. With CASP3 activation, GSDME proteins were cleaved in LV-STING DLBCL cells, indicated by the expression of GSDME-N fragments (Figure 5C).
Interestingly, interactions between necroptosis, apoptosis, and pyroptosis lead to the appearance of PANoptosis. Recent studies demonstrated that CASP8 interacted with RIPK3 and ASC might function as a cell death signaling scaffold to induce PANoptosis [38, 39]. CO-IP assay also revealed that CASP8 directly interacted with RIPK3 and ASC in LV-STING DLBCL cells, which provided the foundation for cell death induction (Figure 5D). Collectively, the above results revealed the importance of STING in PANoptosis induction.
To verify whether PANoptosis concurrently responded to STING activation, we detected the expression of cell death effectors in STING-KO DLBCL cells. As shown in Figure 5E, protein levels of phosphorylated RIPK3 and MLKL were reduced by STING deletion. Besides, cleaved-CASP3 and GSDME-N fragments showed no significance between WT and STING-KO cells (Figure 5F).
As SAMHD1 deficiency led to STING activation, we further explored whether SAMHD1 deficiency induced PANoptosis and, if so, whether STING was the mediator. Consistent with what was observed in STING overexpression, down-regulation of SAMHD1 increased phosphorylated RIPK3, phosphorylated MLKL, cleaved-CASP3, and GSDME-N fragments (Figure 5G). STING deletion conversely decreased the expression of cell death effectors in these SAMHD1-deficient cells (Figure 5G). These data establish that SAMHD1 deficiency promotes STING activation to induce PANoptosis in DLBCL.
DMXAA inhibits DLBCL cell growth by inducing cell death
After showing that STING activation contributed to suppressing DLBCL tumor growth, we tested the anti-tumor effects of STING agonists in these cells. DMXAA is a STING agonist with potential anti-tumor activity [40, 41]. CCK-8 assay showed that DMXAA treatment decreased DLBCL cell viability in a dose-dependent manner (Figure 6A). To explore the mechanisms of DMXAA, we detected phenotypic changes and cell death effectors in DMXAA-treated DLBCL cells. As expected, DMXAA promoted STING expression in DLBCL cells (Figure 6B), consistent with the increase in apoptosis rates and LDH release levels (Figure 6C, D). Immunoblot further verified the activation of cell death effectors, represented by the expression of p-RIPK3, p-MLKL, cleaved-CASP3, and GSDME-N (Figure 6E, F).
To investigate whether the inhibitory effects of DMXAA were dependent on STING, we treated STING-KO DLBCL cells with DMXAA. Genetic deletion of STING significantly decreased the inhibition of DMXAA on cell viability (Figure 6G). As STING expression could be promoted by SAMHD1 deficiency, we further examined the effects of DMXAA on LV-SAMHD1 LY1 cells. Of note, cell viability of SAMHD1-overexpressed cells was significantly suppressed by DMXAA treatment, suggesting that STING activation could overcome SAMHD1-mediated tumor growth (Figure 6H). Together our findings suggest the potential role of STING agonists in anti-DLBCL treatment.
STING activates MLKL, CASP3, and GSDME to induce PANoptosis.A-C. Immunoblot revealed the expression of different cell death effectors in LV-Con and LV-STING DLBCL cells, including the effectors of necroptosis (RIPK3, p-RIPK3, MLKL, and p-MLKL) (A), apoptosis (CASP8, cleaved-CASP8, CASP3, and cleaved-CASP3) (B), and pyroptosis (GSDME-FL and GSDME-N) (C). D. Interactions between CASP8, RIPK3, and ASC in LV-STING DLBCL cells were detected by CO-IP assay. E, F. Immunoblot revealed the expression of cell death effectors in WT and STING-KO DLBCL cells, including the effectors of necroptosis (RIPK3, p-RIPK3, MLKL, and p-MLKL) (E), apoptosis and pyroptosis (CASP3, cleaved-CASP3, GSDME-FL, and GSDME-N) (F). G. Immunoblot revealed the protein levels of p-RIPK3, p-MLKL, cleaved-CASP3, and GSDME-N in LY1 cells transfected with Ctrl, SAMHD1-KD, Ctrl+STING-KO, and SAMHD1-KD+STING-KO. Immunoblot images were the representation of 3 independent experiments.

DMXAA inhibits DLBCL cell growth by inducing cell death. A. LY1 and LY3 cells were treated with the concentration gradients of DMXAA for 24 hours. CCK-8 assay revealed the cell viability and IC50 value in DMXAA-treated cells (LY1 IC50=177μM, LY3 IC50=165μM). B-F. LY1 and LY3 cells were treated with DMSO or 177μM DMXAA for 24 hours. STING expression was measured by immunoblot analysis. Wild-type cells without treatment were the blank control (Blank) (B). Scatter plots (left) and quantitative apoptosis rates (right) were revealed by Annexin V-FITC/PI double staining flow cytometry (C). Supernatant LDH levels were detected by LDH release assay (D). Immunoblot showed the protein levels of cell death effectors in LY1 (E) and LY3 (F) cells. G. WT and STING-KO DLBCL cells were treated with DMSO or 177μM DMXAA for 24 hours. Cell viability was determined by CCK-8 assay. H. LV-Con and LV-SAMHD1 LY1 cells were treated with DMSO or 177μM DMXAA for 24 hours. Cell viability was determined by CCK-8 assay. Vertical bars indicated mean ± SD. P values from unpaired two-tailed t-test (A, C, D, F, G, H). *p<0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.

DMXAA enhances the efficacy of BMS1166 in DLBCL
The potential of STING agonists alone or, more importantly, in combination with immune checkpoint inhibitors draws the great interest of both clinical and pre-clinical research. For example, STING activation can overcome resistance to programmed death 1 (PD-1)/PD-L1 blockade [42]. Given that PD-L1 expression was associated with poor overall survival of DLBCL patients [43], we investigated the efficacy of PD-L1 blockade in DLBCL cells. As shown in Figure 7A, BMS1166, a novel small molecular inhibitor, suppressed cell proliferation in a dose- and time-dependent manner. STING activation was previously demonstrated to boost PD-L1 expression in tumor cells [44]. We also found that the protein levels of PD-L1 were increased by LV-STING and diminished by STING-KO in DLBCL cells (Figure 7B). High levels of PD-L1 were related to high sensitivity to PD-L1 blockade. Notably, BMS1166 treatment significantly reduced the viability of LV-STING DLBCL cells and, to a less degree, decreased the viability of the STING-KO DLBCL cells (Figure 7C, D).
Previous studies illustrated that PD-L1 expression was higher in the activated B-cell (ABC) subtype compared with germinal center B-cell (GCB) subtype [43, 45]. Drug combination investigations were used to address whether the efficacy of PD-L1 blockade could be improved in PD-L1low GCB-like DLBCL. Concentrations below or equal to the IC50 of different agents were used in different regimens (Figure S5A). As shown in Figure 7E, drug combination dramatically decreased the viability of LY1 cells compared to monotherapy. In addition, the CI value was less than 0.8 in a low-dose combination, representing the synergetic effects (Figure 7E).
A cell-derived xenograft (CDX) model was then constructed to investigate the efficacy of the drug combination in vivo (Figure 7F), where DMXAA and BMS1166 functioned at the effective concentrations [46]. Compared with BMS1166 monotherapy, combination treatment significantly suppressed the growth of GCB-like DLBCL cells (Figure 7G, H). Moreover, DMXAA treatment could induce coagulative necrosis in primary tumor lesions (Figure 7H). Collectively, these results support that STING agonist DMXAA enhances the efficacy of PD-L1 blockade in DLBCL.
Discussion
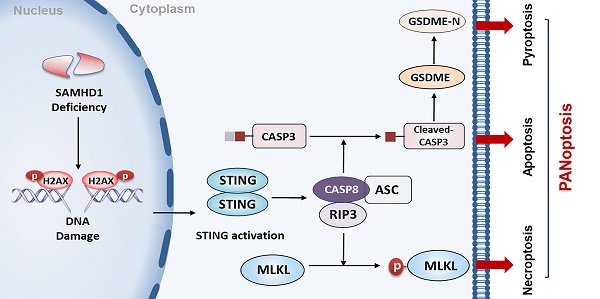
DLBCL, the most common type of lymphoma, is characterized by high levels of genomic instability. It represents a hugely unmet medical need due to our poor understanding of disease development and the lack of good therapeutic targets. Here, we report, for the first time, that STING activation is facilitated by SAMHD1 deficiency and further restricts tumor growth by inducing PANoptosis in DLBCL cells (Figure 8). Additionally, we demonstrate the functions of DMXAA in enhancing the efficacy of anti-PD-L1 treatment, contributing to a potential strategy for DLBCL patients.
In our study, we found that a subset of DLBCL patients with SAMHD1-positive expression was associated with poor prognosis, consistent with the promoting effects of SAMHD1 on tumor growth. SAMHD1 functions in dNTPase-dependent and independent manners [8, 10]. The dNTPase activity of SAMHD1 is essential for restricting virus replication and regulating dNTP pools [47, 48]. However, SAMHD1 overexpression showed no correlation with viral infections in DLBCL patients. According to the enrichment analysis of RNA-seq, we considered that the functional alterations of SAMHD1 might be attributed to cell cycle checkpoints, especially the G2/M DNA damage checkpoint. In particular, cyclin-dependent kinase 1 (CDK1)-induced SAMHD1 phosphorylation might act as a switch that conversed dNTPase-dependent to independent manners, further recruiting nucleic acid exonucleases to restart the stalled DNA double-stranded replication [10]. Therefore, future studies will focus on the post-transcriptional modifications of SAMHD1 in DLBCL. Additional study centers will be included to verify the features of SAMHD1 in DLBCL patients since single-center statistics are limited for characterizing DLBCL as a whole disease.
SAMHD1 has been identified as a negative regulator of innate immunity, as SAMHD1 overexpression prevents the activation of the DNA-sensing pathway [10, 49]. Our data established the crosstalk between SAMHD1 and the cGAS-STING axis in DLBCL. Specifically, SAMHD1 deficiency induced DNA damage and cytosolic dsDNA accumulation, which facilitated activating the cGAS-STING pathway [50, 51]. Of note, the protein expression of STING, rather than cGAS, was increased with SAMHD1 deficiency. A recent study demonstrated that damage repair proteins directly interacted with STING in the process of DNA damage [52]. Therefore, STING activation could be independent of cGAS catalysis. Additionally, our results suggested the specific functions of STING induced by SAMHD1 deficiency. It was well known that STING was associated with cell death regulation and cytokine production [53-55]. Necroptosis, caspase-dependent apoptosis, and pyroptosis were mediated by STING in SAMHD1-deficient DLBCL cells. Caspase activation led to cGAS and IRF3 cleavage, further inhibiting cGAS expression and IFN secretion [56]. Besides, down-regulation of SAMHD1 promoted NF-κB expression to compensate for the silencing of the IFN pathway [38]. Given the importance of cytokines in tumor immune response, future studies will explore the role of STING on cytokine secretion in a natural tumor setting.
DMXAA enhances the efficacy of BMS1166 in DLBCL. A. LY1 and LY3 cells were treated with DMSO or BMS1166 (3, 6, 9, 12μM) for 24 or 48 hours. Cell viability was detected by CCK-8 assay. B. Immunoblot revealed the expression of PD-L1 proteins in LV-Con, LV-STING, WT, and STING-KO DLBCL cells. Wild-type cells without treatment were the blank control (Blank). C, D. DLBCL cells transfected with LV-Con, LV-STING, WT, and STING-KO were treated with DMSO or 9μM BMS1166 for 24 hours. Cell viabilities of LV-STING (C) and STING-KO (D) groups were determined by CCK-8 assay. E. LY1 cells were treated with DMXAA and BMS1166 at a concentration ratio of 88.5:6. Cell viability (upper) and CI values (lower) of different regiments were presented after 24 hours of incubation. F-H. Balb/c nude mice were injected with LY1 cells and randomized into 4 groups. Groups 1-3 were set for monotherapy, where mice were injected with drug-free control or 250μg/ml BMS1166 or 20mg/kg DMXAA every two days. Group 4 was applied for drug combination, where mice were alternately injected with 250μg/ml BMS1166 and 20mg/kg DMXAA every other day (F). Tumor volumes were measured every other day from day 10 (G). Images of tumor bodies were taken on day 18 (H, n=5/group, scale bar=1cm). Immunoblot images were the representation of 3 independent experiments. Vertical bars indicated mean ± SD. P values from unpaired two-tailed t-test (C, D), Welch's one-way ANOVA test with Dunnett's T3 test (E), and Two-way ANOVA analysis with Sidak correction (A, G). *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001.

A proposed model of STING-mediated PANoptosis in DLBCL. SAMHD1 deficiency induced DNA damage to promote STING activation. Activation of STING led to the formation of CASP8/RIPK3/ASC complex, further activating MLKL, CASP3, and GSDME to induce PANoptosis. Specifically, MLKL phosphorylation induced necroptosis. CASP3 cleavage not only induced apoptosis but also cleaved GSDME to induce pyroptosis.

So for, the mechanisms of STING-mediated cell death are undefined in DLBCL. Another important exploration in our study was that STING activation inhibited DLBCL tumor growth by inducing PANoptosis. PANoptosis is a newly discovered PCD with features of necroptosis, apoptosis, and pyroptosis [25, 57]. Increasing evidence highlights the importance of PANoptosis in human cancers, especially in restricting tumorigenesis, regulating immune response, and enhancing chemosensitivity [27, 58, 59]. Constructing a multi-protein complex, known as PANoptosome, is essential for inducing PANoptosis [24]. A recent study illustrated that CASP8/RIPK3/ASC multi-protein complex might work as PANoptosome to activate cell death effectors, which was regulated by NF-κB and tumor necrosis factor-α (TNF-α) [38]. Complex formation further provides conditions for the interactions of different cell death pathways. Necroptosis is a regulated necrosis that requires the activation of RIPK3 and MLKL [60]. As a kind of gasdermin-mediated programmed necrosis, pyroptotic cell death relies on caspase-dependent gasdermin cleavage [61]. Consistent with previous studies, STING activation promotes the formation of CASP8/RIPK3/ASC complex in DLBCL cells, further inducing PANoptosis by activating MLKL, CASP3, and GSDME. These data provide evidence for applying STING agonists in anti-DLBCL treatment.
Several kinds of STING agonists are in clinical trials for solid tumors, such as non-small cell lung carcinoma (NSCLC) and melanoma [18]. Our study highlighted the anti-tumor effects of STING agonist DMXAA in DLBCL. DMXAA monotherapy suppressed cell viability by inducing cell death. In addition, STING activation may enhance the efficacy of immunotherapy in human cancers [62]. Specifically, STING agonists boost anti-tumor immune responses by activating dendritic cells (DCs), natural killer (NK) cells, and IFN-β signaling [62-64]. Although PD-1/PD-L1 immunotherapy has emerged as a promising strategy for DLBCL patients, inadequate treatment responses are presented [65, 66]. We then established the inhibitory effect of BMS1166 on cell proliferation in DLBCL. However, the functions of BMS1166 might be secondary to other non-specific toxic effects [67]. A recent study identifies that activation of STING facilitates overcoming anti-PD-L1 resistance [68]. The drug combination study further supported that targeted activation of STING enhances the efficacy of PD-L1 blockade in DLBCL. On the one hand, STING activation promoted the expression of PD-L1 proteins in PD-L1low GCB-like DLBCL cells, which might enhance the sensitivity to PD-L1 blockade [45]. On the other hand, combination regimens contributed to reducing drug dosage, which was vital for improving the safety of anti-DLBCL treatment. Therefore, STING agonists may emerge as a potential strategy for DLBCL patients, especially those with low levels of PD-L1.
Conclusion
In summary, our study highlights the anti-tumor effects of STING in DLBCL. STING activation responds to SAMHD1 deficiency-induced DNA damage and further induces PANoptosis to suppress tumor growth. Combination of STING agonist and PD-L1 inhibitor enhances the efficacy of PD-L1 blockade in DLBCL, especially in PD-L1low GCB-like DLBCL. These findings provide insights into improving the efficacy of anti-DLBCL treatments.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
Funding
This study was supported by National Natural Science Foundation (No.82270200, No.82170189, No.82070203, No.81800194, No.81770210); Key Research and Development Program of Shandong Province (No.2018CXGC1213); China Postdoctoral Science Foundation (No. 2021T1404223); Translational Research Grant of NCRCH (No.2021WWB02, No.2020ZKMB01); Shandong Provincial Natural Science Foundation (ZR2021YQ51); Taishan Scholars Program of Shandong Province; Shandong Provincial Engineering Research Center of Lymphoma; Academic Promotion Programme of Shandong First Medical University (No. 2019QL018).
Author Contributions
Yiqing Cai and Xiaomin Chen performed the experiments and analyzed the data. Yiqing Cai, Xiaomin Chen and Tiange Lu conducted mice work. Xiaosheng Fang, Mengfei Ding and Zhuoya Yu collected clinical samples. Shunfeng Hu and Jiarui Liu assisted in cell culture. Yiqing Cai and Xiangxiang Zhou edited the paper. Xin Wang and Xiangxiang Zhou revised the manuscript and provided financial support. All authors read and approved the final manuscript.
Availability of Data and Materials
Any additional data used and/or analyzed during the current study are available from the corresponding authors on reasonable request.
Ethics Approval and Consent to Participate
This study was approved by the Medical Ethical Committee of Shandong Provincial Hospital. All samples were obtained with informed consent in accordance with the Declaration of Helsinki. All animal experimental procedures were performed in accordance with the protocols approved by the Institutional Animal Care and Research Advisory Committee of Shandong Provincial Hospital.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A. et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell. 2017;171:481-94 e15
2. Zhu Y, Zhang X, Wei J, Yang C, Tong H, Mai W. et al. Rituximab, lenalidomide and BTK inhibitor as frontline treatment for elderly or unfit patients with diffuse large B-cell lymphoma: a real-world analysis of single center. Exp Hematol Oncol. 2022;11:57
3. Carpio C, Bouabdallah R, Ysebaert L, Sancho JM, Salles G, Cordoba R. et al. Avadomide monotherapy in relapsed/refractory DLBCL: safety, efficacy, and a predictive gene classifier. Blood. 2020;135:996-1007
4. Chu Y, Zhou X, Wang X. Antibody-drug conjugates for the treatment of lymphoma: clinical advances and latest progress. J Hematol Oncol. 2021;14:88
5. Yu Z, Zhou X, Wang X. Metabolic Reprogramming in Hematologic Malignancies: Advances and Clinical Perspectives. Cancer Res. 2022;82:2955-63
6. Mesin L, Ersching J, Victora GD. Germinal Center B Cell Dynamics. Immunity. 2016;45:471-82
7. Restelli V, Lupi M, Chila R, Vagni M, Tarantelli C, Spriano F. et al. DNA Damage Response Inhibitor Combinations Exert Synergistic Antitumor Activity in Aggressive B-Cell Lymphomas. Mol Cancer Ther. 2019;18:1255-64
8. Franzolin E, Pontarin G, Rampazzo C, Miazzi C, Ferraro P, Palumbo E. et al. The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc Natl Acad Sci U S A. 2013;110:14272-7
9. Daddacha W, Koyen AE, Bastien AJ, Head PE, Dhere VR, Nabeta GN. et al. SAMHD1 Promotes DNA End Resection to Facilitate DNA Repair by Homologous Recombination. Cell Rep. 2017;20:1921-35
10. Coquel F, Silva MJ, Techer H, Zadorozhny K, Sharma S, Nieminuszczy J. et al. SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature. 2018;557:57-61
11. Oellerich T, Schneider C, Thomas D, Knecht KM, Buzovetsky O, Kaderali L. et al. Selective inactivation of hypomethylating agents by SAMHD1 provides a rationale for therapeutic stratification in AML. Nat Commun. 2019;10:3475
12. Castellvi M, Felip E, Ezeonwumelu IJ, Badia R, Garcia-Vidal E, Pujantell M. et al. Pharmacological Modulation of SAMHD1 Activity by CDK4/6 Inhibitors Improves Anticancer Therapy. Cancers (Basel). 2020 12
13. Jadersten M, Lilienthal I, Tsesmetzis N, Lourda M, Bengtzen S, Bohlin A. et al. Targeting SAMHD1 with hydroxyurea in first-line cytarabine-based therapy of newly diagnosed acute myeloid leukaemia: Results from the HEAT-AML trial. J Intern Med. 2022
14. Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell. 2014;54:289-96
15. Kwon J, Bakhoum SF. The Cytosolic DNA-Sensing cGAS-STING Pathway in Cancer. Cancer Discov. 2020;10:26-39
16. Lu C, Guan J, Lu S, Jin Q, Rousseau B, Lu T. et al. DNA Sensing in Mismatch Repair-Deficient Tumor Cells Is Essential for Anti-tumor Immunity. Cancer Cell. 2021;39:96-108 e6
17. Hu M, Zhou M, Bao X, Pan D, Jiao M, Liu X. et al. ATM inhibition enhances cancer immunotherapy by promoting mtDNA leakage and cGAS/STING activation. J Clin Invest. 2021 131
18. Le Naour J, Zitvogel L, Galluzzi L, Vacchelli E, Kroemer G. Trial watch: STING agonists in cancer therapy. Oncoimmunology. 2020;9:1777624
19. Zhang R, Kang R, Tang D. The STING1 network regulates autophagy and cell death. Signal Transduct Target Ther. 2021;6:208
20. Tang R, Xu J, Zhang B, Liu J, Liang C, Hua J. et al. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J Hematol Oncol. 2020;13:110
21. Koren E, Fuchs Y. Modes of Regulated Cell Death in Cancer. Cancer Discov. 2021;11:245-65
22. Kist M, Vucic D. Cell death pathways: intricate connections and disease implications. EMBO J. 2021;40:e106700
23. Fritsch M, Gunther SD, Schwarzer R, Albert MC, Schorn F, Werthenbach JP. et al. Caspase-8 is the molecular switch for apoptosis, necroptosis and pyroptosis. Nature. 2019;575:683-7
24. Samir P, Malireddi RKS, Kanneganti TD. The PANoptosome: A Deadly Protein Complex Driving Pyroptosis, Apoptosis, and Necroptosis (PANoptosis). Front Cell Infect Microbiol. 2020;10:238
25. Wang Y, Kanneganti TD. From pyroptosis, apoptosis and necroptosis to PANoptosis: A mechanistic compendium of programmed cell death pathways. Comput Struct Biotechnol J. 2021;19:4641-57
26. Zhu P, Ke ZR, Chen JX, Li SJ, Ma TL, Fan XL. Advances in mechanism and regulation of PANoptosis: Prospects in disease treatment. Front Immunol. 2023;14:1120034
27. Karki R, Sundaram B, Sharma BR, Lee S, Malireddi RKS, Nguyen LN. et al. ADAR1 restricts ZBP1-mediated immune response and PANoptosis to promote tumorigenesis. Cell Rep. 2021;37:109858
28. Messmer MN, Snyder AG, Oberst A. Comparing the effects of different cell death programs in tumor progression and immunotherapy. Cell Death Differ. 2019;26:115-29
29. Huang J, Jiang S, Liang L, He H, Liu Y, Cong L. et al. Analysis of PANoptosis-Related LncRNA-miRNA-mRNA Network Reveals LncRNA SNHG7 Involved in Chemo-Resistance in Colon Adenocarcinoma. Front Oncol. 2022;12:888105
30. Ganapathi KA, Brown LE, Prakash S, Bhargava P. New developments in non-Hodgkin lymphoid malignancies. Pathology. 2021;53:349-66
31. Zhou X, Chen N, Xu H, Zhou X, Wang J, Fang X. et al. Regulation of Hippo-YAP signaling by insulin-like growth factor-1 receptor in the tumorigenesis of diffuse large B-cell lymphoma. J Hematol Oncol. 2020;13:77
32. Liu J, Han Y, Hu S, Cai Y, Yang J, Ren S. et al. Circulating Exosomal MiR-107 Restrains Tumorigenesis in Diffuse Large B-Cell Lymphoma by Targeting 14-3-3eta. Front Cell Dev Biol. 2021;9:667800
33. Zhang X, Bai XC, Chen ZJ. Structures and Mechanisms in the cGAS-STING Innate Immunity Pathway. Immunity. 2020;53:43-53
34. Liu S, Guan W. STING Signaling Promotes Apoptosis, Necrosis, and Cell Death: An Overview and Update. Mediators Inflamm. 2018;2018:1202797
35. Sun L, Wang H, Wang Z, He S, Chen S, Liao D. et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213-27
36. Li X, Li F, Zhang X, Zhang H, Zhao Q, Li M. et al. Caspase-8 auto-cleavage regulates programmed cell death and collaborates with RIPK3/MLKL to prevent lymphopenia. Cell Death Differ. 2022;29:1500-12
37. Lu H, Zhang S, Wu J, Chen M, Cai MC, Fu Y. et al. Molecular Targeted Therapies Elicit Concurrent Apoptotic and GSDME-Dependent Pyroptotic Tumor Cell Death. Clin Cancer Res. 2018;24:6066-77
38. Messaoud-Nacer Y, Culerier E, Rose S, Maillet I, Rouxel N, Briault S. et al. STING agonist diABZI induces PANoptosis and DNA mediated acute respiratory distress syndrome (ARDS). Cell Death Dis. 2022;13:269
39. Wang Y, Pandian N, Han JH, Sundaram B, Lee S, Karki R. et al. Single cell analysis of PANoptosome cell death complexes through an expansion microscopy method. Cell Mol Life Sci. 2022;79:531
40. Curran E, Chen X, Corrales L, Kline DE, Dubensky TW Jr, Duttagupta P. et al. STING Pathway Activation Stimulates Potent Immunity against Acute Myeloid Leukemia. Cell Rep. 2016;15:2357-66
41. Liang J, Wang H, Ding W, Huang J, Zhou X, Wang H. et al. Nanoparticle-enhanced chemo-immunotherapy to trigger robust antitumor immunity. Sci Adv. 2020;6:eabc3646
42. Fu J, Kanne DB, Leong M, Glickman LH, McWhirter SM, Lemmens E. et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci Transl Med. 2015;7:283ra52
43. Kiyasu J, Miyoshi H, Hirata A, Arakawa F, Ichikawa A, Niino D. et al. Expression of programmed cell death ligand 1 is associated with poor overall survival in patients with diffuse large B-cell lymphoma. Blood. 2015;126:2193-201
44. Zhou L, Xu Q, Huang L, Jin J, Zuo X, Zhang Q. et al. Low-dose carboplatin reprograms tumor immune microenvironment through STING signaling pathway and synergizes with PD-1 inhibitors in lung cancer. Cancer Lett. 2021;500:163-71
45. Hu LY, Xu XL, Rao HL, Chen J, Lai RC, Huang HQ. et al. Expression and clinical value of programmed cell death-ligand 1 (PD-L1) in diffuse large B cell lymphoma: a retrospective study. Chin J Cancer. 2017;36:94
46. Seshadri M, Spernyak JA, Mazurchuk R, Camacho SH, Oseroff AR, Cheney RT. et al. Tumor vascular response to photodynamic therapy and the antivascular agent 5,6-dimethylxanthenone-4-acetic acid: implications for combination therapy. Clin Cancer Res. 2005;11:4241-50
47. Zhang K, Lv DW, Li R. Conserved Herpesvirus Protein Kinases Target SAMHD1 to Facilitate Virus Replication. Cell Rep. 2019;28:449-59 e5
48. Hu J, Qiao M, Chen Y, Tang H, Zhang W, Tang D. et al. Cyclin E2-CDK2 mediates SAMHD1 phosphorylation to abrogate its restriction of HBV replication in hepatoma cells. FEBS Lett. 2018;592:1893-904
49. Chen S, Bonifati S, Qin Z, St Gelais C, Kodigepalli KM, Barrett BS. et al. SAMHD1 suppresses innate immune responses to viral infections and inflammatory stimuli by inhibiting the NF-kappaB and interferon pathways. Proc Natl Acad Sci U S A. 2018;115:E3798-E807
50. Ahn J, Ruiz P, Barber GN. Intrinsic self-DNA triggers inflammatory disease dependent on STING. J Immunol. 2014;193:4634-42
51. Xu Y, Nowsheen S, Deng M. DNA Repair Deficiency Regulates Immunity Response in Cancers: Molecular Mechanism and Approaches for Combining Immunotherapy. Cancers (Basel). 2023 15
52. Dunphy G, Flannery SM, Almine JF, Connolly DJ, Paulus C, Jonsson KL. et al. Non-canonical Activation of the DNA Sensing Adaptor STING by ATM and IFI16 Mediates NF-kappaB Signaling after Nuclear DNA Damage. Mol Cell. 2018;71:745-60 e5
53. Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788-92
54. Martin GR, Salazar Arcila C, Hallihan LJ, Scheidl-Yee T, Jirik FR. Inducible generalized activation of hSTING-N154S expression in mice leads to lethal hypercytokinemia: a model for "cytokine storm". J Leukoc Biol. 2023;113:326-33
55. Zheng W, Liu A, Xia N, Chen N, Meurens F, Zhu J. How the Innate Immune DNA Sensing cGAS-STING Pathway Is Involved in Apoptosis. Int J Mol Sci. 2023 24
56. Ning X, Wang Y, Jing M, Sha M, Lv M, Gao P. et al. Apoptotic Caspases Suppress Type I Interferon Production via the Cleavage of cGAS, MAVS, and IRF3. Mol Cell. 2019;74:19-31 e7
57. Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature. 2015;517:311-20
58. Liu J, Hong M, Li Y, Chen D, Wu Y, Hu Y. Programmed Cell Death Tunes Tumor Immunity. Front Immunol. 2022;13:847345
59. Lin JF, Hu PS, Wang YY, Tan YT, Yu K, Liao K. et al. Phosphorylated NFS1 weakens oxaliplatin-based chemosensitivity of colorectal cancer by preventing PANoptosis. Signal Transduct Target Ther. 2022;7:54
60. Garcia LR, Tenev T, Newman R, Haich RO, Liccardi G, John SW. et al. Ubiquitylation of MLKL at lysine 219 positively regulates necroptosis-induced tissue injury and pathogen clearance. Nat Commun. 2021;12:3364
61. Kovacs SB, Miao EA. Gasdermins: Effectors of Pyroptosis. Trends Cell Biol. 2017;27:673-84
62. Yi M, Niu M, Wu Y, Ge H, Jiao D, Zhu S. et al. Combination of oral STING agonist MSA-2 and anti-TGF-beta/PD-L1 bispecific antibody YM101: a novel immune cocktail therapy for non-inflamed tumors. J Hematol Oncol. 2022;15:142
63. Nakamura T, Sato T, Endo R, Sasaki S, Takahashi N, Sato Y. et al. STING agonist loaded lipid nanoparticles overcome anti-PD-1 resistance in melanoma lung metastasis via NK cell activation. J Immunother Cancer. 2021 9
64. Yin M, Hu J, Yuan Z, Luo G, Yao J, Wang R. et al. STING agonist enhances the efficacy of programmed death-ligand 1 monoclonal antibody in breast cancer immunotherapy by activating the interferon-beta signalling pathway. Cell Cycle. 2022;21:767-79
65. Wang C, Shi F, Liu Y, Zhang Y, Dong L, Li X. et al. Anti-PD-1 antibodies as a salvage therapy for patients with diffuse large B cell lymphoma who progressed/relapsed after CART19/20 therapy. J Hematol Oncol. 2021;14:106
66. Dai M, Liu M, Yang H, Kucuk C, You H. New insights into epigenetic regulation of resistance to PD-1/PD-L1 blockade cancer immunotherapy: mechanisms and therapeutic opportunities. Exp Hematol Oncol. 2022;11:101
67. Zhou L, Chai F, He Y, Zhou Z, Guo S, Li P. et al. Homodimerized cytoplasmic domain of PD-L1 regulates its complex glycosylation in living cells. Commun Biol. 2022;5:887
68. Zhu L, Gao H, Huang S, Cao T, Zhai X, Hu J. et al. Combination of STING agonist and CXCR3 antagonist disrupts immune tolerance to overcome anti-PD-L1 resistance in lung adenocarcinoma under oxidative stress. Gene. 2022;851:146962
Author contact
![]() Corresponding authors: Xin Wang, M.D., Ph.D. Director & Professor, Department of Hematology, Shandong Provincial Hospital, Shandong University, No.324, Jingwu Road, Jinan, Shandong 250021, China; Tel: 0086-531-68776358 (B); 0086-13156012606 (M); Fax: 0086-531-87061197 (B); Email: xinwedu.cn. Xiangxiang Zhou, M.D., Ph.D. Professor, Department of Hematology, Shandong Provincial Hospital Affiliated to Shandong First Medical University, No.324, Jingwu Road, Jinan, Shandong 250021, China; Tel: 0086-531-68776358(B); 0086-15866695595(M); Email: xiangxiangzhouedu.cn.
Corresponding authors: Xin Wang, M.D., Ph.D. Director & Professor, Department of Hematology, Shandong Provincial Hospital, Shandong University, No.324, Jingwu Road, Jinan, Shandong 250021, China; Tel: 0086-531-68776358 (B); 0086-13156012606 (M); Fax: 0086-531-87061197 (B); Email: xinwedu.cn. Xiangxiang Zhou, M.D., Ph.D. Professor, Department of Hematology, Shandong Provincial Hospital Affiliated to Shandong First Medical University, No.324, Jingwu Road, Jinan, Shandong 250021, China; Tel: 0086-531-68776358(B); 0086-15866695595(M); Email: xiangxiangzhouedu.cn.
Received 2023-4-13
Accepted 2023-8-11
Published 2023-8-28
